8A8E
 
 | | PPSA C terminal octahedral structure | | Descriptor: | Phosphoenolpyruvate synthase | | Authors: | Song, W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase
To Be Published
|
|
4Z0C
 
 | | Crystal structure of TLR13-ssRNA13 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-R(P*AP*CP*GP*GP*AP*AP*AP*GP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2015-03-26 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13
Nat.Struct.Mol.Biol., 22, 2015
|
|
5HYX
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-SER-ASN-PRO-GLY-HIS-HIS-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-01-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth
Cell Res., 26, 2016
|
|
5HZ1
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-ARG-ALA-LYS-HIS-HIS-PRO-HYP-LYS-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HZ0
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-LYS-PRO-ARG-HIS-HIS-PRO-HYP-ARG-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-04-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HZ3
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGFR5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-PRO-LYS-PRO-SER-THR-ARG-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Plant Receptor
To Be Published
|
|
9EPU
 
 | | Crystal structure of the human CDKL5 kinase domain with compound CAF-382 | | Descriptor: | Cyclin-dependent kinase-like 5, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2024-03-20 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human CDKL5 kinase domain with compound CAF-382
To Be Published
|
|
7XOZ
 
 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
9ETY
 
 | | KEAP1 in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(2~{R})-2,4-dimethyl-3-oxidanylidene-piperazin-1-yl]-2,3-dihydro-1,3-thiazol-4-yl]sulfonyl]benzenecarbonitrile, Kelch-like ECH-associated protein 1, ... | | Authors: | Richardson, W, Bullock, A.N, Rothweiler, E.M, Manning, C.E, Sweeney, M.N, Chalk, R, Huber, K.V.M. | | Deposit date: | 2024-03-27 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Covalent Inhibitors of KEAP1 with Exquisite Selectivity.
J.Med.Chem., 67, 2024
|
|
9ETW
 
 | | BTB domain of KLHL26 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 26 | | Authors: | Richardson, W, Bullock, A.N, Rothweiler, E.M, Manning, C.E, Sweeney, M.N, Chalk, R, Huber, K.V.M. | | Deposit date: | 2024-03-27 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Covalent Inhibitors of KEAP1 with Exquisite Selectivity.
J.Med.Chem., 67, 2024
|
|
9ETX
 
 | | KEAP1 BTB in complex with compound 23 | | Descriptor: | (3R)-4-[4-(2-chlorophenyl)sulfonyl-1,3-thiazol-5-yl]-1,3-dimethyl-piperazin-2-one, Kelch-like ECH-associated protein 1 | | Authors: | Richardson, W, Bullock, A.N, Rothweiler, E.M, Manning, C.E, Sweeney, M.N, Chalk, R, Huber, K.V.M. | | Deposit date: | 2024-03-27 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Covalent Inhibitors of KEAP1 with Exquisite Selectivity.
J.Med.Chem., 67, 2024
|
|
8CIE
 
 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
6ACC
 
 | |
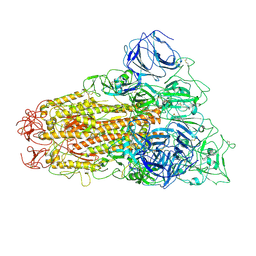
6ACD
 
 | |
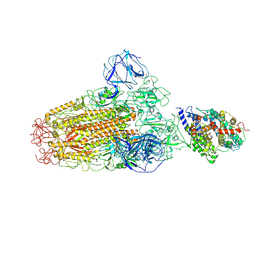
6ACG
 
 | |
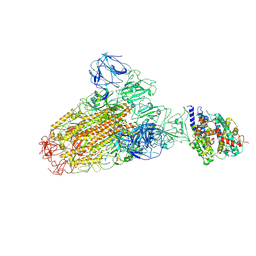
6ACJ
 
 | |
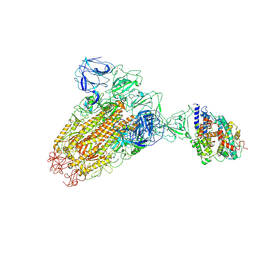
6ACK
 
 | |
3SKS
 
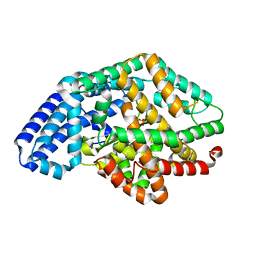 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
7GH2
 
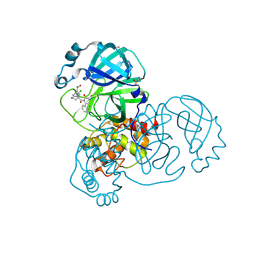 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHE
 
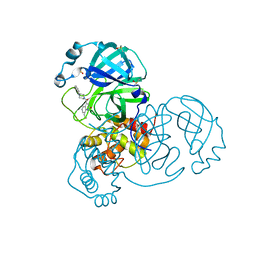 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-916a2c5a-4 (Mpro-x3303) | | Descriptor: | 3C-like proteinase, 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH4
 
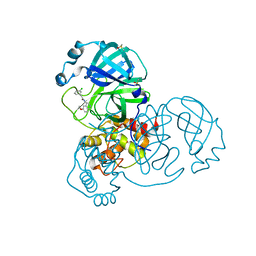 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | Descriptor: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-4 (Mpro-x10566) | | Descriptor: | 2-(3-cyclopropylphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCT
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-LEF-c49414a7-1 (Mpro-x10756) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with SIM-SYN-f15aaa3a-1 (Mpro-x3348) | | Descriptor: | 1-[4-(diphenylmethyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
