3DAD
 
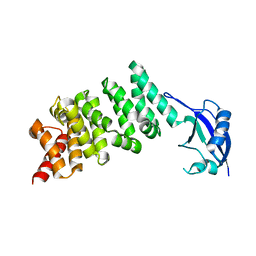 | | Crystal structure of the N-terminal regulatory domains of the formin FHOD1 | | Descriptor: | FH1/FH2 domain-containing protein 1 | | Authors: | Schulte, A, Stolp, B, Schonichen, A, Pylypenko, O, Rak, A, Fackler, O.T, Geyer, M. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Human Formin FHOD1 Contains a Bipartite Structure of FH3 and GTPase-Binding Domains Required for Activation.
Structure, 16, 2008
|
|
3REB
 
 | |
3REA
 
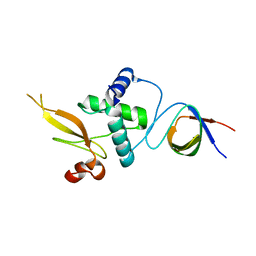 | |
2PK2
 
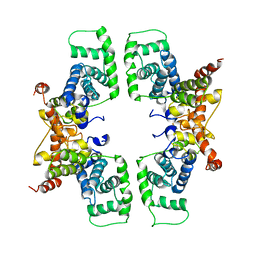 | | Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat | | Descriptor: | Cyclin-T1, Protein Tat | | Authors: | Anand, K, Schulte, A, Fujinaga, K, Scheffzek, K, Geyer, M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Cyclin Box Structure of the P-TEFb Subunit Cyclin T1 Derived from a Fusion Complex with EIAV Tat.
J.Mol.Biol., 370, 2007
|
|
3RBB
 
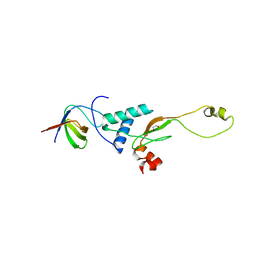 | | HIV-1 NEF protein in complex with engineered HCK SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein Nef, Tyrosine-protein kinase HCK | | Authors: | Horenkamp, F.A, Schulte, A, Weyand, M, Geyer, M. | | Deposit date: | 2011-03-29 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation of the Dileucine-Based Sorting Motif in HIV-1 Nef Revealed by Intermolecular Domain Assembly.
Traffic, 12, 2011
|
|
2HYB
 
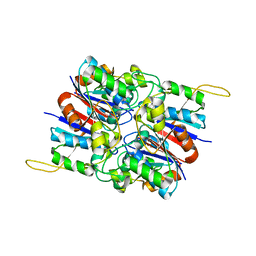 | | Crystal Structure of Hexameric DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexameric DsrEFH
To be Published
|
|
2HY5
 
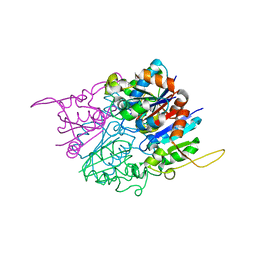 | | Crystal structure of DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of DsrEFH
To be Published
|
|
2GD7
 
 | |
1YX3
 
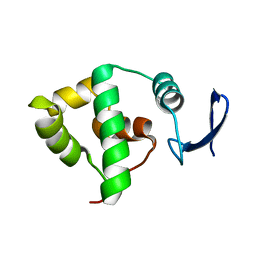 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
7WHG
 
 | |
7WHF
 
 | | Heimdallarchaeota gelsolin (2DGel) bound to rabbit actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2021-12-30 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evidence for the emergence of a calcium-regulated actin cytoskeleton prior to eukaryogenesis
Commun Biol, 5, 2022
|
|
4P5S
 
 | | Structure of reduced W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
4P5R
 
 | | Structure of oxidized W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, SODIUM ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
2W3S
 
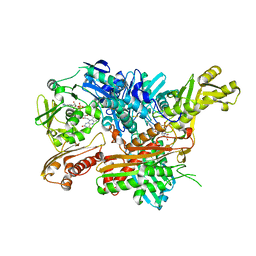 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in Complex with Xanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
5MDR
 
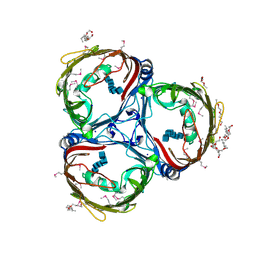 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
5MDO
 
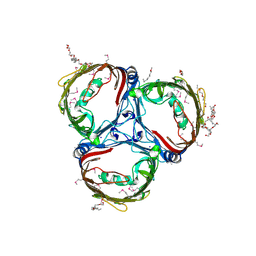 | |
5MDP
 
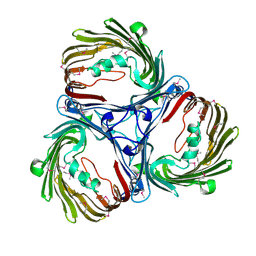 | |
5MDQ
 
 | |
5MDS
 
 | |
2W54
 
 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W3R
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W55
 
 | | Crystal Structure of Xanthine Dehydrogenase (E232Q variant) from Rhodobacter capsulatus in Complex with Hypoxanthine | | Descriptor: | BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2W2H
 
 | |
8QX6
 
 | | Novel laminarin-binding CBM X584 | | Descriptor: | PKD domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Zuehlke, M.K, Jeudy, A, Czjzek, M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ.Microbiol., 26, 2024
|
|
6HHN
 
 | |
