1D7Y
 
 | | CRYSTAL STRUCTURE OF NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4 | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsui, Y. | | Deposit date: | 1999-10-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
1F3P
 
 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
4I86
 
 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | Descriptor: | Cellulose synthase 1 | | Authors: | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
3DMI
 
 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
8IN4
 
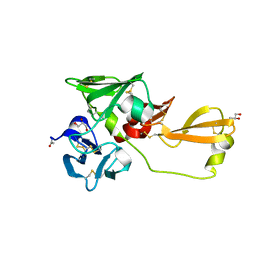 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
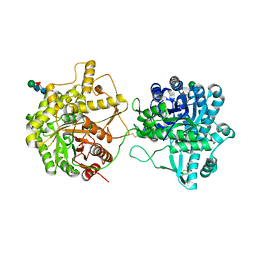 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
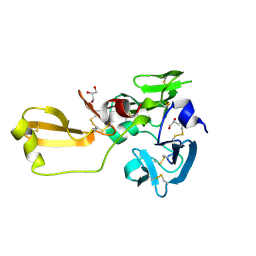 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
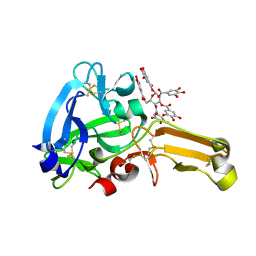 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
5AYE
 
 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate and beta-(1,4)-mannobiose | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AYD
 
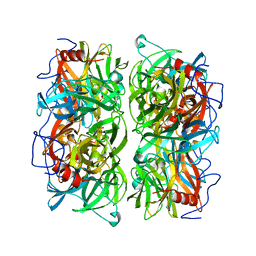 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AY9
 
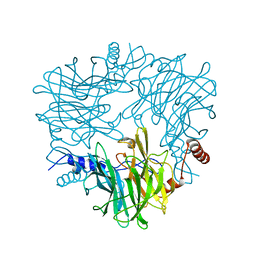 | |
5AYC
 
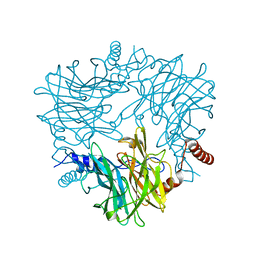 | | Crystal structure of Ruminococcus albus 4-O-beta-D-mannosyl-D-glucose phosphorylase (RaMP1) in complexes with sulfate and 4-O-beta-D-mannosyl-D-glucose | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, SULFATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
1ULI
 
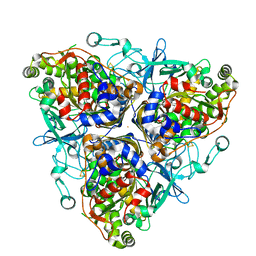 | | Biphenyl dioxygenase (BphA1A2) derived from Rhodococcus sp. strain RHA1 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, biphenyl dioxygenase large subunit, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1ULJ
 
 | | Biphenyl dioxygenase (BphA1A2) in complex with the substrate | | Descriptor: | BIPHENYL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
