5O4P
 
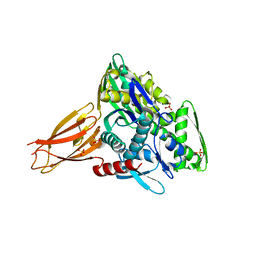 | | Crystal structure of AMPylated GRP78 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Yan, Y, Chen, R, Ron, D, Read, R. | | Deposit date: | 2017-05-30 | | Release date: | 2017-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5MLG
 
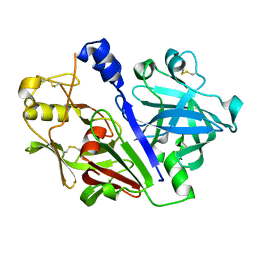 | | Crystal structure of rat prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Yan, Y, Read, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat prorenin
To Be Published
|
|
5MKT
 
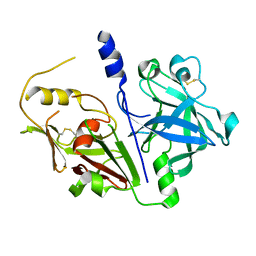 | | Crystal structure of mouse prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin-1 | | Authors: | Yan, Y, Read, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of mouse prorenin
To Be Published
|
|
3CEY
 
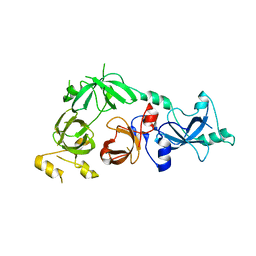 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
6MRL
 
 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
6MRM
 
 | | Red Clover Necrotic Mosaic Virus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
4CSP
 
 | | Structure of the F306C mutant of nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Leferink, N.G.H, Antonyuk, S.V, Houwman, J.A, Scrutton, N.S, REady, R, Hasnain, S.S. | | Deposit date: | 2014-03-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of Residues Remote from the Catalytic Centre on Enzyme Catalysis of Copper Nitrite Reductase.
Nat.Commun., 5, 2014
|
|
4CSZ
 
 | | STRUCTURE OF F306C MUTANT OF NITRITE REDUCTASE FROM Achromobacter XYLOSOXIDANS WITH NITRITE BOUND | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Leferink, N.G.H, Antonyuk, S.V, Houwman, J.A, Scrutton, N.S, REady, R, Hasnain, S.S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of Residues Remote from the Catalytic Centre on Enzyme Catalysis of Copper Nitrite Reductase.
Nat.Commun., 5, 2014
|
|
