2N8J
 
 | |
5TP6
 
 | |
5TP5
 
 | |
2LL6
 
 | | Solution NMR structure of CaM bound to iNOS CaM binding domain peptide | | Descriptor: | Calmodulin, Nitric oxide synthase, inducible | | Authors: | Piazza, M, Futrega, K, Spratt, D.E, Dieckmann, T, Guillemette, J.G. | | Deposit date: | 2011-10-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides: Effects of a Phosphomimetic CaM Mutation.
Biochemistry, 51, 2012
|
|
2MG5
 
 | |
2LL7
 
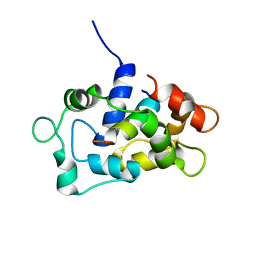 | | Solution NMR structure of CaM bound to the eNOS CaM binding domain peptide | | Descriptor: | Calmodulin, Nitric oxide synthase, endothelial | | Authors: | Piazza, M, Futrega, K, Spratt, D.E, Guillemette, J.G, Dieckmann, T. | | Deposit date: | 2011-10-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides: Effects of a Phosphomimetic CaM Mutation.
Biochemistry, 51, 2012
|
|
6UZJ
 
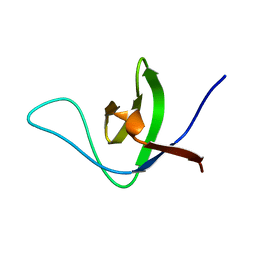 | | NMR structure of the HACS1 SH3 domain | | Descriptor: | SAM domain-containing protein SAMSN-1 | | Authors: | Donaldson, L.W. | | Deposit date: | 2019-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HACS1 signaling adaptor protein recognizes a motif in the paired immunoglobulin receptor B cytoplasmic domain.
Commun Biol, 3, 2020
|
|
7UCP
 
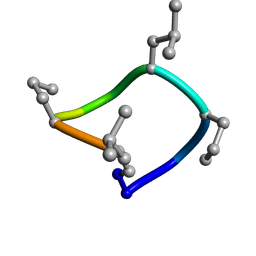 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
7UZL
 
 | |
6WHO
 
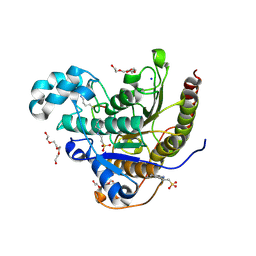 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
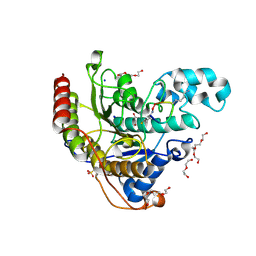 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WSJ
 
 | |
6WHQ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
