3MIN
 
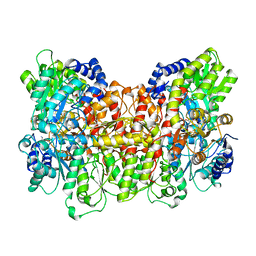 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
1LRV
 
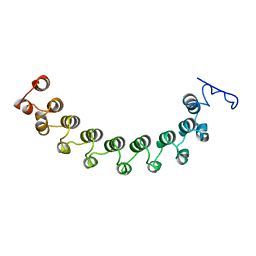 | |
1FEH
 
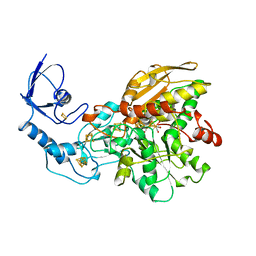 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
2MIN
 
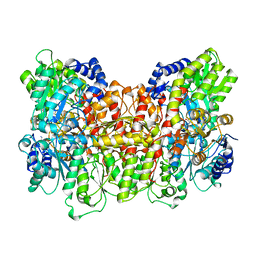 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
3K1A
 
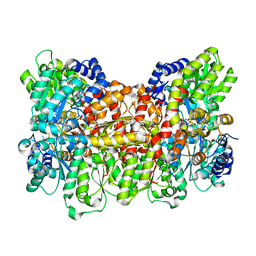 | | Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70Ile MoFe protein variant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Peters, J.W, Sarma, R, Barney, B.M, Keable, S, Seefeldt, L.C, Dean, D.R. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70(Ile) MoFe protein variant
J.Inorg.Biochem., 104, 2010
|
|
5JFC
 
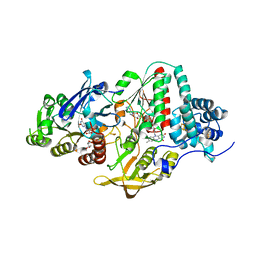 | | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, King, P.W, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
5M45
 
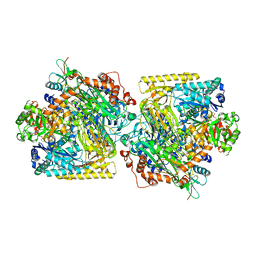 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
4XPI
 
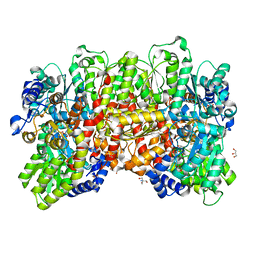 | | Fe protein independent substrate reduction by nitrogenase variants altered in intramolecular electron transfer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Danyal, K, Rasmusen, A.J, Keable, S.M, Shaw, S, Zadvornyy, O, Duval, S, Dean, D.R, Raugei, S, Peters, J.W, Seefeldt, L.C. | | Deposit date: | 2015-01-17 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fe protein-independent substrate reduction by nitrogenase MoFe protein variants.
Biochemistry, 54, 2015
|
|
6VWE
 
 | |
3C8Y
 
 | | 1.39 Angstrom crystal structure of Fe-only hydrogenase | | Descriptor: | 2 IRON/2 SULFUR/3 CARBONYL/2 CYANIDE/WATER/METHYLETHER CLUSTER, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Pandey, A.S, Lemon, B.J, Peters, J.W. | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Dithiomethylether as a ligand in the hydrogenase h-cluster.
J.Am.Chem.Soc., 130, 2008
|
|
6N6P
 
 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
3LX4
 
 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
6NAC
 
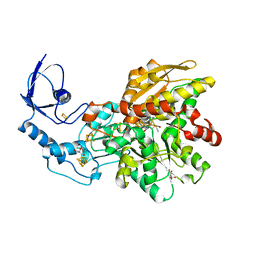 | | Crystal structure of [FeFe]-hydrogenase I (CpI) solved with single pulse free electron laser data | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Cohen, A.E, Davidson, C.M, Zadvornyy, O.A, Keable, S.M, Lyubimov, A.Y, Song, J, McPhillips, S.E, Soltis, S.M, Peters, J.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6N59
 
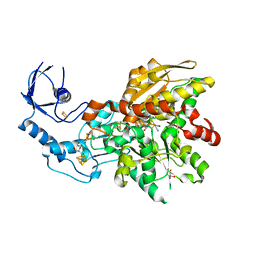 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
4YWO
 
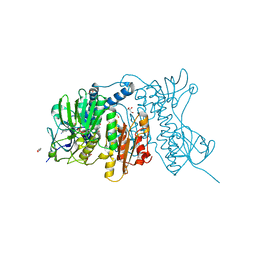 | | Mercuric reductase from Metallosphaera sedula | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase | | Authors: | Artz, J.H, Zadvornyy, O.A, White, S, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula.
Front Bioeng Biotechnol, 3, 2015
|
|
4Z1Y
 
 | |
4Z17
 
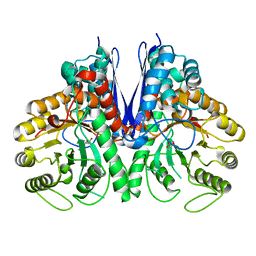 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
4YWS
 
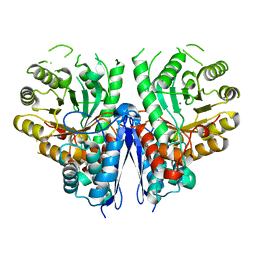 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
5JCA
 
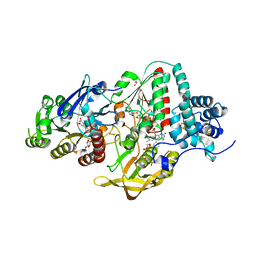 | | NADP(H) bound NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
2C3C
 
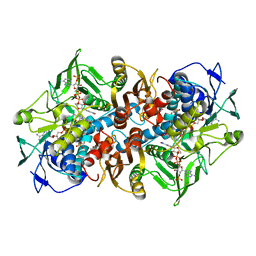 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2C8V
 
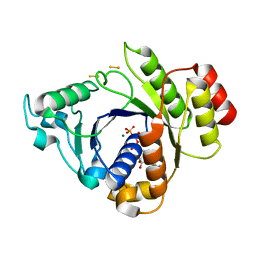 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2CFC
 
 | | structural basis for stereo selectivity in the (R)- and (S)- hydroxypropylethane thiosulfonate dehydrogenases | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 2-(R)-HYDROXYPROPYL-COM DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Krishnakumar, A.M, Nocek, B.P, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2006-02-19 | | Release date: | 2006-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Stereoselectivity in the (R)-and (S)-Hydroxypropylthioethanesulfonate Dehydrogenases.
Biochemistry, 45, 2006
|
|
6BBL
 
 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
6CDK
 
 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
