7MMW
 
 | |
7MMT
 
 | |
7MMU
 
 | |
7MMV
 
 | |
7MMQ
 
 | |
7MMP
 
 | |
7MMS
 
 | |
7MMR
 
 | |
6EBP
 
 | |
6EBO
 
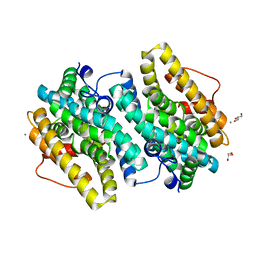 | | Crystal Structure of the Class Ie Ribonucleotide Reductase Beta Subunit from Aerococcus urinae in Unactivated Form | | Descriptor: | CALCIUM ION, GLYCEROL, Ribonucleoside-diphosphate reductase, ... | | Authors: | Palowitch, G.M, Alapati, R.B, Boal, A.K. | | Deposit date: | 2018-08-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Metal-free class Ie ribonucleotide reductase from pathogens initiates catalysis with a tyrosine-derived dihydroxyphenylalanine radical.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EBQ
 
 | |
6EBZ
 
 | |
6DQX
 
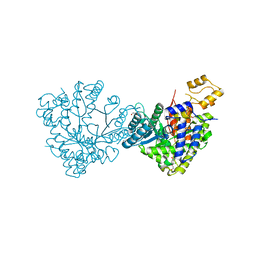 | | Actinobacillus ureae class Id ribonucleotide reductase alpha subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McBride, M.J, Palowitch, G.M, Boal, A.K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis.
Biochemistry, 58, 2019
|
|
6DQW
 
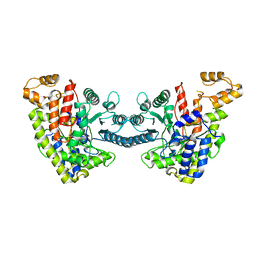 | |
8CVD
 
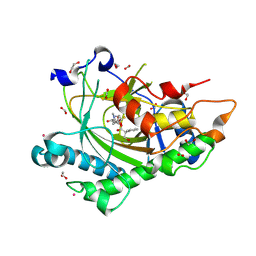 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVF
 
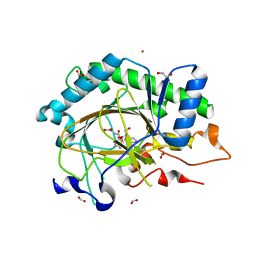 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVH
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVA
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CV9
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVG
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CV8
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVC
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
