3WX5
 
 | |
3WYD
 
 | |
3X17
 
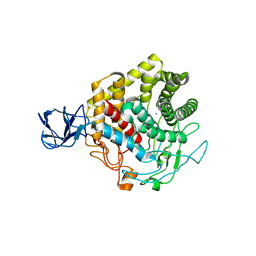 | | Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase | | Descriptor: | CALCIUM ION, Endoglucanase, ZINC ION | | Authors: | Okano, H, Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, activity, and stability of metagenome-derived glycoside hydrolase family 9 endoglucanase with an N-terminal Ig-like domain.
Protein Sci., 24, 2015
|
|
2ZXC
 
 | | Ceramidase complexed with C2 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
5X7K
 
 | |
1UAW
 
 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
2MST
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2RS2
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5NEN
 
 | |
