6QUB
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum in complex with galactose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUC
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUD
 
 | | 2-deoxy-galactose reaction intermediate of a Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, CALCIUM ION | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6EHO
 
 | | Dimer of the Sortilin Vps10p domain at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thirup, S.S, Quistgaard, E.H, Januliene, D, Andersen, J.L, Nielsen, J.A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Acidic Environment Induces Dimerization and Ligand Binding Site Collapse in the Vps10p Domain of Sortilin.
Structure, 25, 2017
|
|
7PG2
 
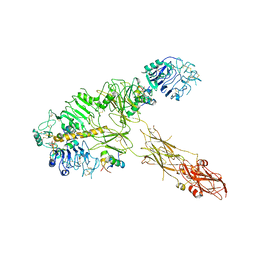 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG0
 
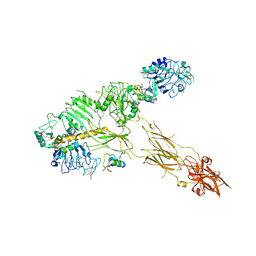 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG4
 
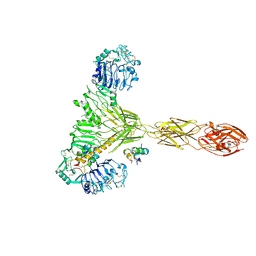 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG3
 
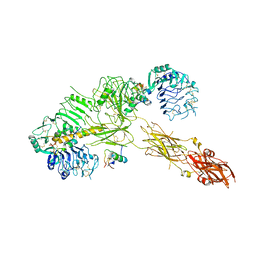 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
