2I76
 
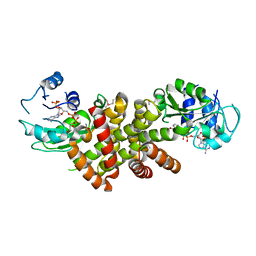 | | Crystal structure of protein TM1727 from Thermotoga maritima | | Descriptor: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
2IPQ
 
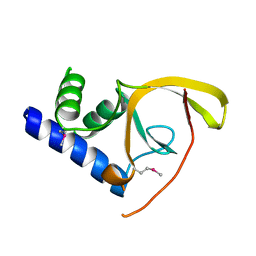 | | Crystal structure of C-terminal domain of Salmonella Enterica protein STY4665, PFAM DUF1528 | | Descriptor: | Hypothetical protein STY4665 | | Authors: | Ramagopal, U.A, Bonanno, J.B, Gilmore, J, Toro, R, Bain, K.T, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-terminal domain of Hypothetical protein STY4665
To be Published
|
|
1VHY
 
 | |
2L5L
 
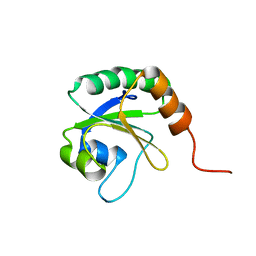 | | Solution Structure of Thioredoxin from Bacteroides Vulgatus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Thioredoxin from Bacteroides Vulgatus
To be Published
|
|
2L5O
 
 | | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis | | Descriptor: | Putative thioredoxin | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Putative Thioredoxin from Neisseria meningitidis
To be Published
|
|
2L57
 
 | | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens
To be Published
|
|
1VQW
 
 | | Crystal structure of a protein with similarity to flavin-containing monooxygenases and to mammalian dimethylalanine monooxygenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-05 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1LW5
 
 | |
1M0W
 
 | | Yeast Glutathione Synthase Bound to gamma-glutamyl-cysteine, AMP-PNP and 2 Magnesium Ions | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-14 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Conformational Changes in the Catalytic Cycle of Glutathione Synthase
Structure, 10, 2002
|
|
1M0T
 
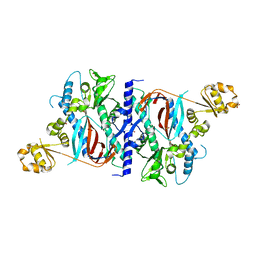 | |
1LX7
 
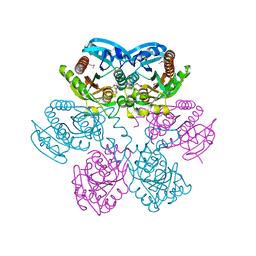 | | Structure of E. coli uridine phosphorylase at 2.0A | | Descriptor: | uridine phosphorylase | | Authors: | Burling, T, Buglino, J.A, Kniewel, R, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-04 | | Release date: | 2002-06-12 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli uridine phosphorylase at 2.0 A.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3BPD
 
 | |
3CAW
 
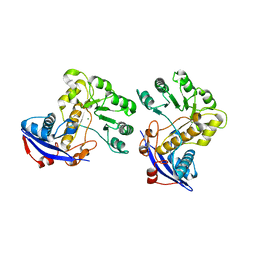 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3BQ9
 
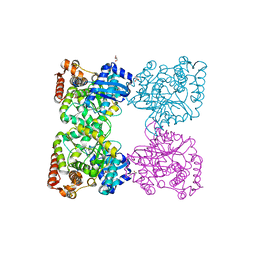 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | Descriptor: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
3BVC
 
 | | Crystal structure of uncharacterized protein Ism_01780 from Roseovarius nubinhibens ISM | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Uncharacterized protein Ism_01780 | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Rutter, M, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-06 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an uncharacterized protein Ism_01780 from Roseovarius nubinhibens.
To be Published
|
|
3CE2
 
 | | Crystal structure of putative peptidase from Chlamydophila abortus | | Descriptor: | Putative peptidase, ZINC ION | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Eberle, M, Maletic, M, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative peptidase from Chlamydophila abortus.
To be Published
|
|
3C8C
 
 | | Crystal structure of Mcp_N and cache domains of methyl-accepting chemotaxis protein from Vibrio cholerae | | Descriptor: | ALANINE, MAGNESIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Hu, S, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-19 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae.
To be Published
|
|
3C9F
 
 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
3CB3
 
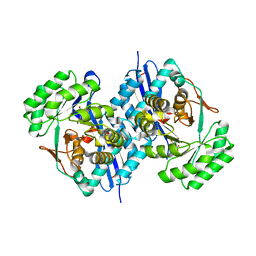 | | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate | | Descriptor: | L-GLUCARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate.
To be Published
|
|
3CBW
 
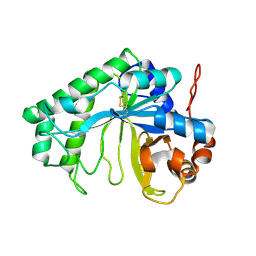 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
3CT2
 
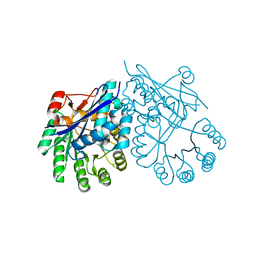 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
3COK
 
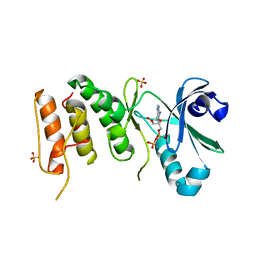 | | Crystal structure of PLK4 kinase | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Atwell, S, Burley, S.K, Houle, A, Leon, B, Pelletier, L.A, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of PLK4 kinase.
To be Published
|
|
3CDX
 
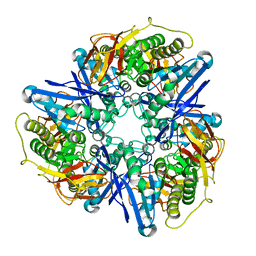 | | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides | | Descriptor: | CALCIUM ION, Succinylglutamatedesuccinylase/aspartoacylase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Patterson, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides.
To be Published
|
|
3CIH
 
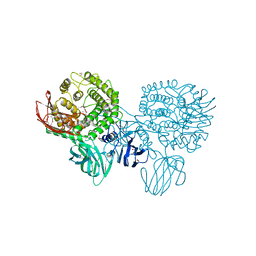 | | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative alpha-rhamnosidase | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-11 | | Release date: | 2008-04-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron.
To be Published
|
|
3C9G
 
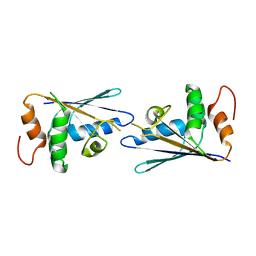 | |
