2RKN
 
 | | X-ray structure of the self-defense and signaling protein DIR1 from Arabidopsis taliana | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, DIR1 protein, ZINC ION | | Authors: | Lascombe, M.B, Prange, T, Buhot, N, Marion, D, Bakan, B, Lamb, C. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein.
Protein Sci., 17, 2008
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | Descriptor: | RNA polymerase alpha subunit | | Authors: | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | Deposit date: | 2003-10-06 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
1TOF
 
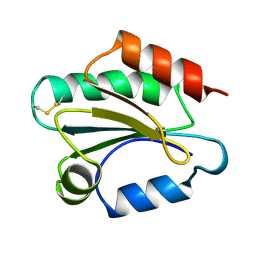 | | THIOREDOXIN H (OXIDIZED FORM), NMR, 23 STRUCTURES | | Descriptor: | THIOREDOXIN H | | Authors: | Mittard, V, Blackledge, M.J, Stein, M, Jacquot, J.-P, Marion, D, Lancelin, J.-M. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii.
Eur.J.Biochem., 243, 1997
|
|
1GH1
 
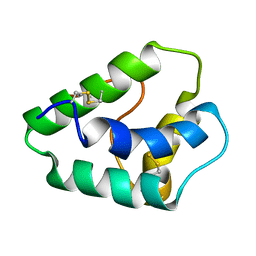 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
1AIW
 
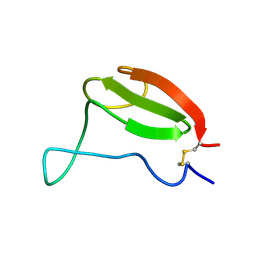 | | NMR STRUCTURES OF THE CELLULOSE-BINDING DOMAIN OF THE ENDOGLUCANASE Z FROM ERWINIA CHRYSANTHEMI, 23 STRUCTURES | | Descriptor: | ENDOGLUCANASE Z | | Authors: | Brun, E, Moriaud, F, Gans, P, Blackledge, M.J, Barras, F, Marion, D. | | Deposit date: | 1997-04-30 | | Release date: | 1998-05-06 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi.
Biochemistry, 36, 1997
|
|
2DVH
 
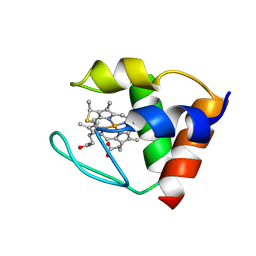 | | THE Y64A MUTANT OF CYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH, NMR, 39 STRUCTURES | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Sebban-Kreuzer, C, Blackledge, M.J, Dolla, A, Marion, D, Guerlesquin, F. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
3GSH
 
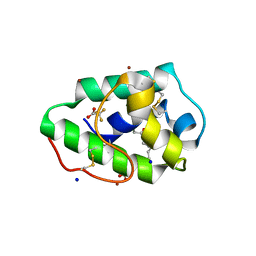 | | Three-dimensional structure of a post translational modified barley LTP1 | | Descriptor: | (12E)-10-oxooctadec-12-enoic acid, Non-specific lipid-transfer protein 1, SODIUM ION, ... | | Authors: | Lascombe, M.B, Prange, T, Bakan, B, Marion, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of oxylipin-conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid-binding proteins.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
1IMT
 
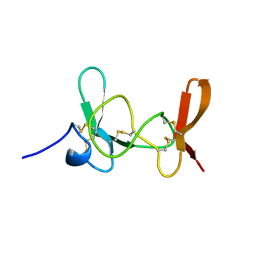 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
1I5V
 
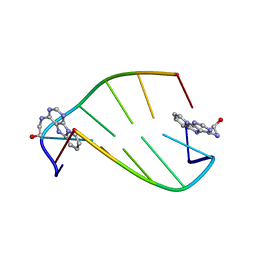 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
1DVH
 
 | | STRUCTURE AND DYNAMICS OF FERROCYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS STUDIED BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | CYTOCHROME C553, HEME C | | Authors: | Blackledge, M.J, Medvedeva, S, Poncin, M, Guerlesquin, F, Bruschi, M, Marion, D. | | Deposit date: | 1995-02-24 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
1GKS
 
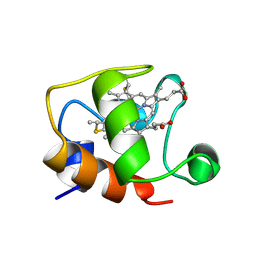 | | ECTOTHIORHODOSPIRA HALOPHILA CYTOCHROME C551 (REDUCED), NMR, 37 STRUCTURES | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bersch, B, Blackledge, M.J, Meyer, T.E, Marion, D. | | Deposit date: | 1996-07-16 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Ectothiorhodospira halophila ferrocytochrome c551: solution structure and comparison with bacterial cytochromes c.
J.Mol.Biol., 264, 1996
|
|
1AFH
 
 | | LIPID TRANSFER PROTEIN FROM MAIZE SEEDLINGS, NMR, 15 STRUCTURES | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gomar, J, Petit, M.C, Sodano, P, Sy, D, Marion, D, Kader, J.C, Vovelle, F, Ptak, M. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds.
Protein Sci., 5, 1996
|
|
1TFS
 
 | | NMR AND RESTRAINED MOLECULAR DYNAMICS STUDY OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF TOXIN FS2, A SPECIFIC BLOCKER OF THE L-TYPE CALCIUM CHANNEL, ISOLATED FROM BLACK MAMBA VENOM | | Descriptor: | TOXIN FS2 | | Authors: | Albrand, J.-P, Blackledge, M.J, Pascaud, F, Hollecker, M, Marion, D. | | Deposit date: | 1995-01-26 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR and restrained molecular dynamics study of the three-dimensional solution structure of toxin FS2, a specific blocker of the L-type calcium channel, isolated from black mamba venom.
Biochemistry, 34, 1995
|
|
1S62
 
 | | Solution structure of the Escherichia coli TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Deprez, C, Blanchard, L, Simorre, J.-P, Gavioli, M, Guerlesquin, F, Lazdunski, C, Lloubes, R, Marion, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain.
J.Mol.Biol., 346, 2005
|
|
1C2N
 
 | | CYTOCHROME C2, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Cordier, F, Caffrey, M.S, Brutscher, B, Cusanovich, M.A, Marion, D, Blackledge, M. | | Deposit date: | 1998-04-27 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, rotational diffusion anisotropy and local backbone dynamics of Rhodobacter capsulatus cytochrome c2.
J.Mol.Biol., 281, 1998
|
|
1APQ
 
 | | STRUCTURE OF THE EGF-LIKE MODULE OF HUMAN C1R, NMR, 19 STRUCTURES | | Descriptor: | COMPLEMENT PROTEASE C1R | | Authors: | Bersch, B, Hernandez, J.-F, Marion, D, Arlaud, G.J. | | Deposit date: | 1997-07-22 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor (EGF)-like module of human complement protease C1r, an atypical member of the EGF family.
Biochemistry, 37, 1998
|
|
1EKY
 
 | |
1CZ2
 
 | | SOLUTION STRUCTURE OF WHEAT NS-LTP COMPLEXED WITH PROSTAGLANDIN B2. | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PROSTAGLANDIN B2 | | Authors: | Tassin-Moindrot, S, Caille, A, Douliez, J.-P, Marion, D, Vovelle, F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2.
Eur.J.Biochem., 267, 2000
|
|
1DBY
 
 | | NMR STRUCTURES OF CHLOROPLAST THIOREDOXIN M CH2 FROM THE GREEN ALGA CHLAMYDOMONAS REINHARDTII | | Descriptor: | CHLOROPLAST THIOREDOXIN M CH2 | | Authors: | Lancelin, J.-M, Guilhaudis, L, Krimm, I, Blackledge, M.J, Marion, D. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thioredoxin m from the green alga Chlamydomonas reinhardtii.
Proteins, 41, 2000
|
|
6I1B
 
 | |
7I1B
 
 | |
1DEM
 
 | |
1DEN
 
 | |
1BWO
 
 | |
