6I19
 
 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin h1 | | Descriptor: | Thioredoxin H-type | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.378 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
6I1C
 
 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin f2 | | Descriptor: | thioredoxin f2 | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
7ZUV
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic sedoheptulose-1,7-bisphosphatase in reducing conditions | | Descriptor: | FBPase domain-containing protein, SULFATE ION | | Authors: | Le Moigne, T, Robert, G.Q, Lemaire, S.D, Henri, J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 194, 2024
|
|
6H7G
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | Descriptor: | Phosphoribulokinase, chloroplastic, SULFATE ION | | Authors: | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4MKN
 
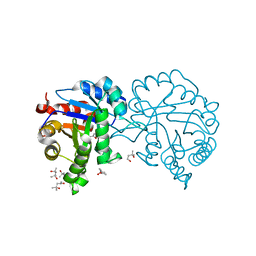 | | Crystal structure of chloroplastic triosephosphate isomerase from Chlamydomonas reinhardtii at 1.1 A of resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Triosephosphate isomerase | | Authors: | Fermani, S, Sciabolini, C, Zaffagnini, M, Lemaire, S.D. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structure and Redox Properties of Chloroplastic Triosephosphate Isomerase from Chlamydomonas reinhardtii.
Mol Plant, 7, 2014
|
|
7ZY7
 
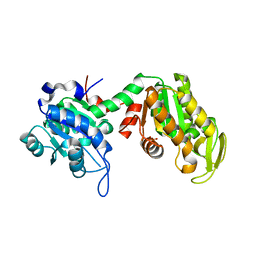 | |
7ZQK
 
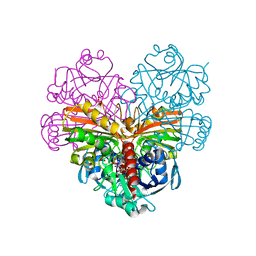 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ4
 
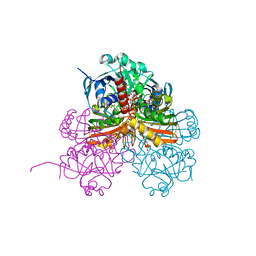 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ3
 
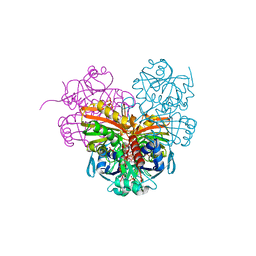 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
6Q6V
 
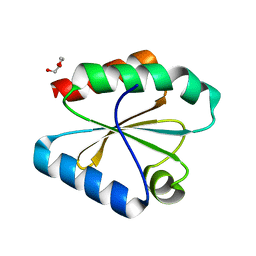 | |
6Q47
 
 | |
6Q6T
 
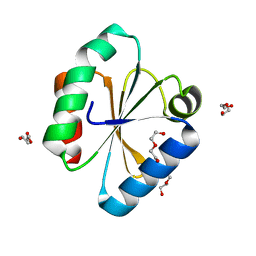 | |
6Q46
 
 | |
6Q6U
 
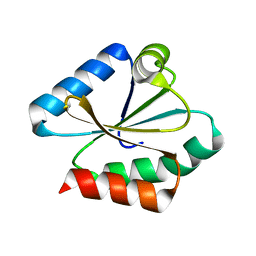 | |
6ZXT
 
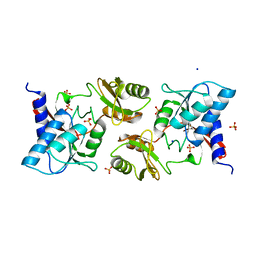 | | High resolution crystal structure of chloroplastic ribose-5-phosphate isomerase from Chlamydomonas reinhardtii | | Descriptor: | Ribose-5-phosphate isomerase, SODIUM ION, SULFATE ION | | Authors: | Le Moigne, T, Crozet, P, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Crystal Structure of Chloroplastic Ribose-5-Phosphate Isomerase from Chlamydomonas reinhardtii -An Enzyme Involved in the Photosynthetic Calvin-Benson Cycle.
Int J Mol Sci, 21, 2020
|
|
7AAS
 
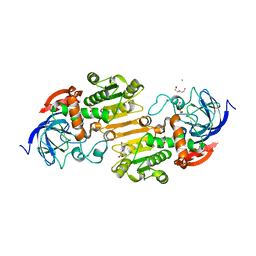 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7AV7
 
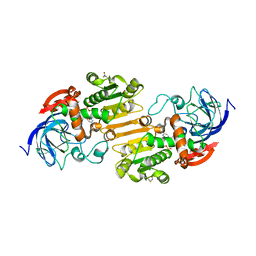 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
7ASW
 
 | | Crystal structure of chloroplastic thioredoxin z defines a novel type-specific target recognition | | Descriptor: | Thioredoxin-related protein CITRX | | Authors: | Le Moigne, T, Gurrieri, L, Crozet, P, Marchand, C.H, Zaffagnini, M, Sparla, F, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-10-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Crystal structure of chloroplastic thioredoxin z defines a type-specific target recognition.
Plant J., 107, 2021
|
|
7B1W
 
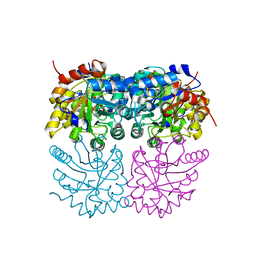 | | Crystal structure of plastidial ribulose epimerase RPE1 from the model alga Chlamydomonas reinhardtii | | Descriptor: | Ribulose-phosphate 3-epimerase, ZINC ION | | Authors: | Henri, J, Zaffagnini, M, Tedesco, D, Crozet, P, Lemaire, S.D. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 2023
|
|
7B2O
 
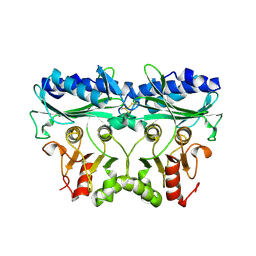 | |
7B2N
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic Fructose bisphosphate aldolase | | Descriptor: | CHLORIDE ION, Fructose-bisphosphate aldolase 1, chloroplastic, ... | | Authors: | Le Moigne, T, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of chloroplast fructose-1,6-bisphosphate aldolase from the green alga Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2022
|
|
7AAU
 
 | | Crystal structure of nitrosoglutathione reductase from Chlamydomonas reinhardtii in complex with NAD+ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
6QUN
 
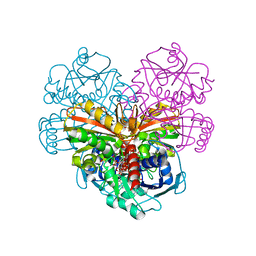 | | Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QUQ
 
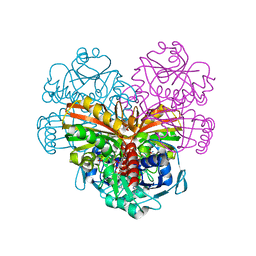 | | Crystal structure of glutathionylated glycolytic glyceraldehyde-3- phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1) | | Descriptor: | GLUTATHIONE, Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Trost, P. | | Deposit date: | 2019-02-28 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5ND5
 
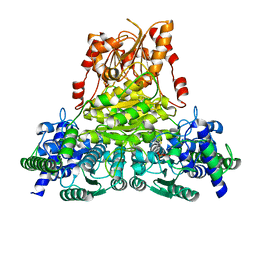 | | Crystal structure of transketolase from Chlamydomonas reinhardtii in complex with TPP and Mg2+ | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Fermani, S, Zaffagnini, M, Francia, F, Pasquini, M. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the magnesium-dependent activation of transketolase from Chlamydomonas reinhardtii.
Biochim. Biophys. Acta, 1861, 2017
|
|
