7SXP
 
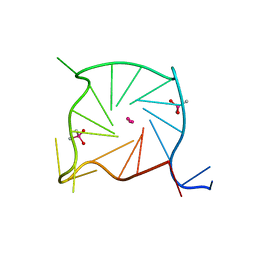 | |
7E9I
 
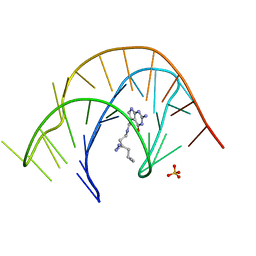 | | Crystal structure of a class I PreQ1 riboswitch aptamer (wild-type) complexed with a cognate ligand-derived photoaffinity probe | | Descriptor: | 2-azanyl-5-[[2-(3-but-3-ynyl-1,2-diazirin-3-yl)ethylamino]methyl]-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one, 33-mer RNA, SULFATE ION | | Authors: | Numata, T, Schneekloth, J.S. | | Deposit date: | 2021-03-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites.
Nat Commun, 12, 2021
|
|
7E9E
 
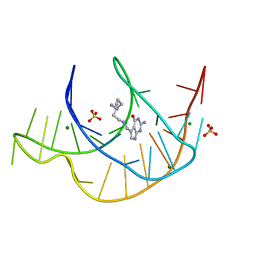 | | Crystal structure of a class I PreQ1 riboswitch aptamer (ab13-14) complexed with a cognate ligand-derived photoaffinity probe | | Descriptor: | 2-azanyl-5-[[2-(3-but-3-ynyl-1,2-diazirin-3-yl)ethylamino]methyl]-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one, 33-mer RNA, MAGNESIUM ION, ... | | Authors: | Numata, T, Schneekloth, J.S. | | Deposit date: | 2021-03-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites.
Nat Commun, 12, 2021
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7PBH
 
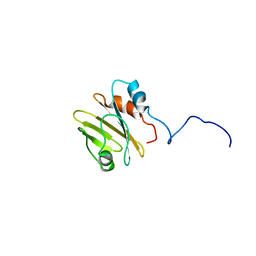 | |
