1KY9
 
 | | Crystal Structure of DegP (HtrA) | | Descriptor: | PROTEASE DO | | Authors: | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2002-02-04 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
3CS0
 
 | | Crystal structure of DegP24 | | Descriptor: | Periplasmic serine endoprotease DegP, pentapeptide | | Authors: | Krojer, T, Sawa, J, Schaefer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2008-04-08 | | Release date: | 2008-05-27 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
6SE4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
3MH4
 
 | |
3MH5
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH6
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH7
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | 5-mer peptide, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (2.961 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
5MQ4
 
 | | Crystal Structure of the leucine zipper of human PRKCBP1 | | Descriptor: | Protein kinase C-binding protein 1, SULFATE ION, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Picaud, S, Newman, J, Tallant, C, Heroven, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the leucine zipper of human PRKCBP1
To Be Published
|
|
7AV9
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
7AV8
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7BBP
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
3OOY
 
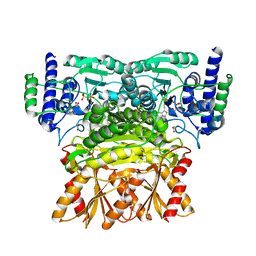 | | Crystal structure of human Transketolase (TKT) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Krojer, T, Krysztofinska, E, Guo, K, Pilka, E, Kochan, G, Chaikuad, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Transketolase (TKT)
To be Published
|
|
5CLT
 
 | | Crystal structure of human glycogen branching enzyme (GBE1) in complex with acarbose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krojer, T, Froese, D.S, Goubin, S, Strain-Damerell, C, Mahajan, P, Burgess-Brown, N, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human glycogen branching enzyme (GBE1) in complex with acarbose
To be published
|
|
5CLW
 
 | | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krojer, T, Froese, D.S, Goubin, S, Strain-Damerell, C, Mahajan, P, Burgess-Brown, N, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose
To be published
|
|
4D6S
 
 | | crystal structure of human JMJD2D in complex with N-OXALYLGLYCINE and bound 5,6-Dimethylbenzimidazole | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Vollmar, M, Bradley, A, Crawley, L, Szykowska, A, Burgess-Brown, N, Gileadi, C, Johansson, C, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Human Jmjd2D in Complex with N-Oxalylglycine and Bound 5,6- Dimethylbenzimidazole
To be Published
|
|
4D6R
 
 | | crystal structure of human JMJD2D in complex with N-OXALYLGLYCINE and bound o-toluenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 4D, N-OXALYLGLYCINE, ... | | Authors: | Krojer, T, Vollmar, M, Bradley, A, Crawley, L, Szykowska, A, Burgess-Brown, N, Gileadi, C, Johansson, C, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystal Structure of Human Jmjd2D in Complex with N-Oxalylglycine and Bound O-Toluenesulfonamide
To be Published
|
|
4D6Q
 
 | | crystal structure of human JMJD2D in complex with 2,4-PDCA | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 4D, NICKEL (II) ION, ... | | Authors: | Krojer, T, Vollmar, M, Bradley, A, Crawley, L, Szykowska, A, Burgess-Brown, N, Gileadi, C, Johansson, C, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | Crystal Structure of Human Jmjd2D in Complex with 2,4-Pdca
To be Published
|
|
4DIP
 
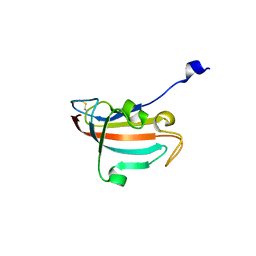 | | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14 | | Descriptor: | PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase FKBP14, SODIUM ION | | Authors: | Krojer, T, Kiyani, W, Goubin, S, Muniz, J.R.C, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Oppermann, U, Zschocke, J, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14
To be Published
|
|
5S8M
 
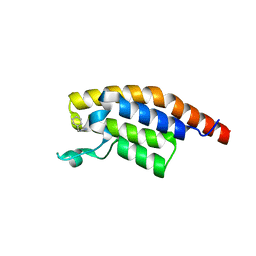 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N11511a (space group C2) | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8G
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N00804d (space group C2) | | Descriptor: | 1,3-benzothiazole-6-carboxylic acid, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8K
 
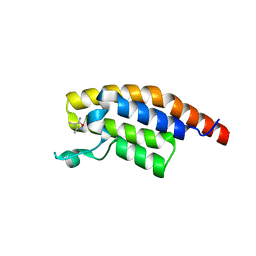 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N01225c (space group C2) | | Descriptor: | 3,4-dimethyl-1,2-oxazol-5-amine, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8J
 
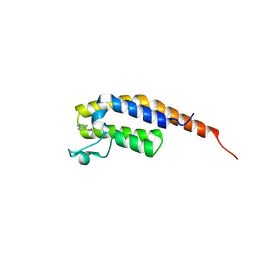 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N01207d (space group C2) | | Descriptor: | 5-chloro-N,1-dimethyl-1H-pyrazole-4-carboxamide, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8Q
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with FMO3D000185a (space group P212121) | | Descriptor: | (3R)-3-methyl-1,2,3,4-tetrahydro-5H-1,4-benzodiazepin-5-one, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8C
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N00322e (space group C2) | | Descriptor: | (1-methylbenzotriazol-5-yl)methanol, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | XChem group deposition
To Be Published
|
|
