4AXG
 
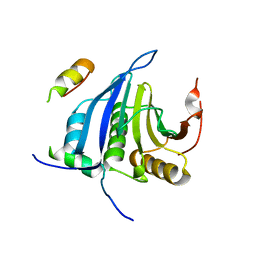 | | Structure of eIF4E-Cup complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, PROTEIN CUP | | Authors: | Kinkelin, K, Veith, K, Gruenwald, M, Bono, F. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Minimal Eif4E-Cup Complex Revelas a General Mechanism of Eif4E Regulation in Translational Repression
RNA, 18, 2012
|
|
4BY7
 
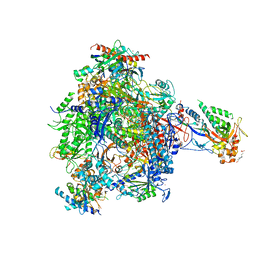 | | elongating RNA Polymerase II-Bye1 TLD complex | | Descriptor: | , 5'-D(*DAP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*DTP)-3', 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BY1
 
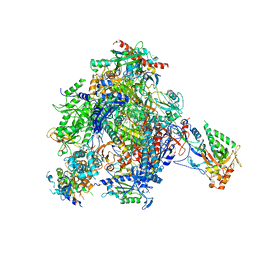 | | elongating RNA Polymerase II-Bye1 TLD complex soaked with AMPCPP | | Descriptor: | 5'-D(*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP *TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*AP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BXZ
 
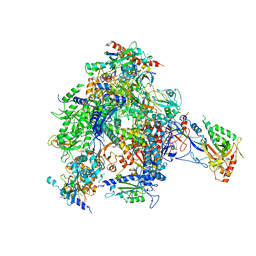 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BXX
 
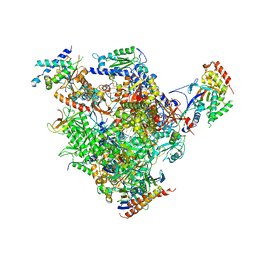 | | Arrested RNA polymerase II-Bye1 complex | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*AP*CP*CP*TP*GP *GP*TP*GP* BRUP*TP*GP*CP*TP*CP*TP*AP*AP*DC)-3', 5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP)-3', 5'-D(*GP*AP*GP*GP*TP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5MEC
 
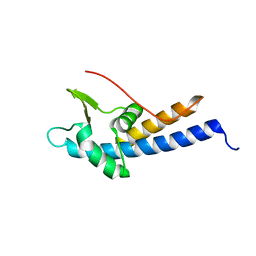 | |
5MEA
 
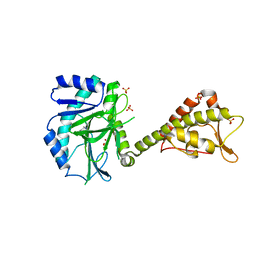 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 2. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5ME9
 
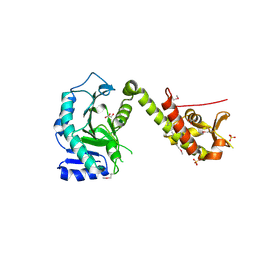 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | Descriptor: | Cell division cycle protein CDT1, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
