2Q7G
 
 | | Pyrrolysine tRNA Synthetase bound to a pyrrolysine analogue (cyc) and ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q7E
 
 | | The structure of pyrrolysyl-tRNA synthetase bound to an ATP analogue | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q7H
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3N1O
 
 | | Crystal structure of IhhN | | Descriptor: | CALCIUM ION, Indian hedgehog protein, ZINC ION | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
3N1G
 
 | | Crystal structure of DhhN bound to BOCFn3 | | Descriptor: | Brother of CDO, CALCIUM ION, Desert hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
3N1R
 
 | | Crystal Structure of ShhN | | Descriptor: | CALCIUM ION, Sonic hedgehog protein, ZINC ION | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of ShhN
TO BE PUBLISHED
|
|
3N1P
 
 | | Crystal Structure of IhhN bound to BOCFn3 | | Descriptor: | Brother of CDO, CALCIUM ION, Indian hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
3N1Q
 
 | | Crystal Structure of DhhN bound to CDOFn3 | | Descriptor: | CALCIUM ION, Cell adhesion molecule-related/down-regulated by oncogenes, Desert hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
3N1F
 
 | | Crystal Structure of IhhN bound to CDOFn3 | | Descriptor: | CALCIUM ION, Cell adhesion molecule-related/down-regulated by oncogenes, Indian hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
3N1M
 
 | | Crystal Structure of IhhN bound to BOCFn3 | | Descriptor: | Brother of CDO, CALCIUM ION, Indian hedgehog protein, ... | | Authors: | Kavran, J.M, Leahy, D.J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner.
J.Biol.Chem., 285, 2010
|
|
2ZIM
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Steitz, T.A, Kavran, J.M. | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4PGF
 
 | | The structure of mono-acetylated SAHH | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
4PFJ
 
 | | The structure of bi-acetylated SAHH | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
2QA4
 
 | |
6BN1
 
 | | Salvador Hippo SARAH domain complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, Scaffold protein salvador, ... | | Authors: | Cairns, L, Kavran, J.M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salvador has an extended SARAH domain that mediates binding to Hippo kinase.
J. Biol. Chem., 293, 2018
|
|
6TYK
 
 | |
4Y6C
 
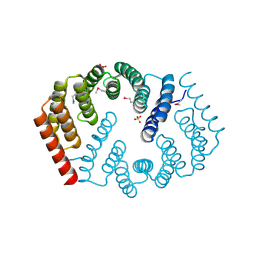 | | Q17M crystal structure of Podosopora anserina putative kinesin light chain nearly identical TPR-like repeats | | Descriptor: | ANSERINA PUTATIVE KINESIN LIGHT chain, SULFATE ION | | Authors: | Marold, J.D, Kavran, J.M, Bowman, G.D, Barrick, D. | | Deposit date: | 2015-02-12 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | A Naturally Occurring Repeat Protein with High Internal Sequence Identity Defines a New Class of TPR-like Proteins.
Structure, 23, 2015
|
|
4Y6W
 
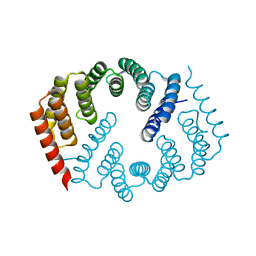 | |
6Q1B
 
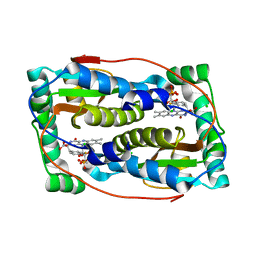 | |
6PZ0
 
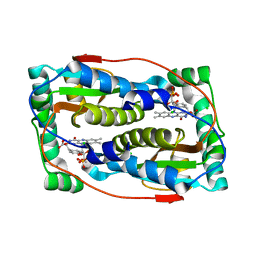 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and L-Tyrosine | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, TYROSINE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-07-31 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
6Q1L
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
5KRD
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and 2-iodophenol (2IP) | | Descriptor: | 2-iodanylphenol, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
5KO8
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN and mono-iodotyrosine (I-Tyr) | | Descriptor: | 3-IODO-TYROSINE, FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S.E. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
5KO7
 
 | | Crystal structure of haliscomenobacter hydrossis iodotyrosine deiodinase (IYD) bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Ingavat, N, Kavran, J.M, Sun, Z, Rokita, S. | | Deposit date: | 2016-06-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Active Site Binding Is Not Sufficient for Reductive Deiodination by Iodotyrosine Deiodinase.
Biochemistry, 56, 2017
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
