200D
 
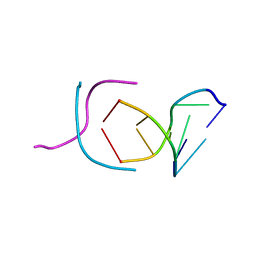 | | STABLE LOOP IN THE CRYSTAL STRUCTURE OF THE INTERCALATED FOUR-STRANDED CYTOSINE-RICH METAZOAN TELOMERE | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*C)-3') | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1D59
 
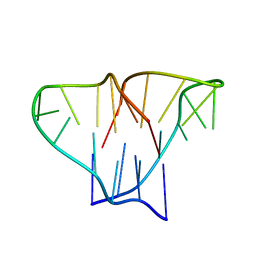 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1992-02-25 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
2MF9
 
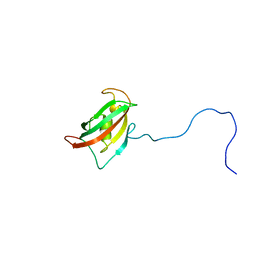 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
1REG
 
 | |
2K21
 
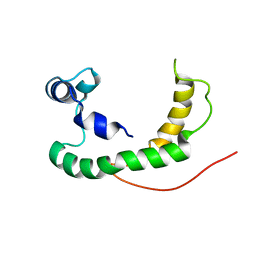 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
6PXS
 
 | | Crystal structure of iminodiacetate oxidase (IdaA) from Chelativorans sp. BNC1 | | Descriptor: | FAD dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Jun, S.Y, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural and biochemical characterization of iminodiacetate oxidase from Chelativorans sp. BNC1.
Mol.Microbiol., 112, 2019
|
|
3K88
 
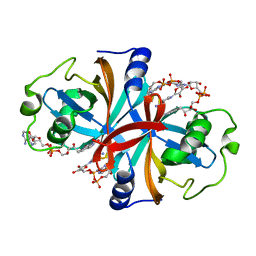 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD, NADH complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kang, C, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
9B1R
 
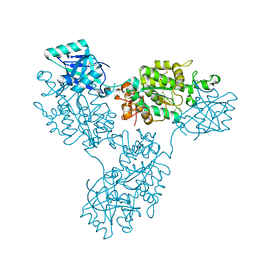 | | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1 | | Authors: | Park, H, Youn, B, Park, D.J, Puthanveettil, S.V, Kang, C. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 (VSR1) from Arabidopsis thaliana.
Sci Rep, 14, 2024
|
|
2J3H
 
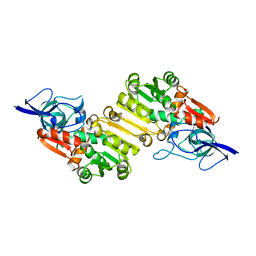 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
5KN1
 
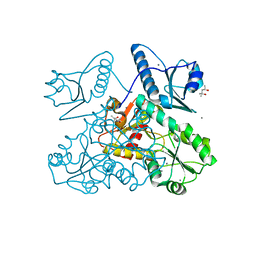 | | Recombinant bovine skeletal calsequestrin, high-Ca2+ form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
4LBH
 
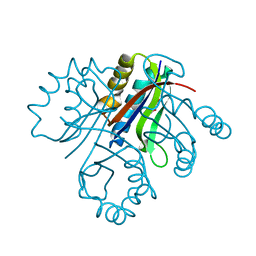 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4LBI
 
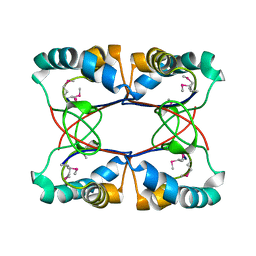 | |
5KN0
 
 | | Native bovine skeletal calsequestrin, low-Ca2+ form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
5KN2
 
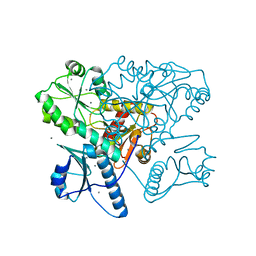 | | Native bovine skeletal calsequestrin, high-Ca2+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Calsequestrin | | Authors: | Lewis, K.M, Byrd, S.S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
5KN3
 
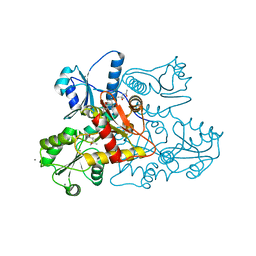 | | Recombinant bovine skeletal calsequestrin, low-Ca2+ form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
6P7K
 
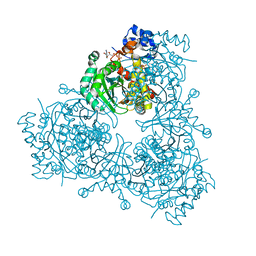 | | Structure of HMG-CoA reductase from Burkholderia cenocepacia | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A reductase, ADENOSINE-5'-DIPHOSPHATE, COENZYME A | | Authors: | Walker, A.M, Peacock, R.B, Hicks, C.W, Dewing, S.M, Lewis, K.M, Abboud, J, Stewart, S.W.A, Kang, C, Watson, J.M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural and Functional Characterization of Dynamic Oligomerization in Burkholderia cenocepacia HMG-CoA Reductase.
Biochemistry, 58, 2019
|
|
5CRG
 
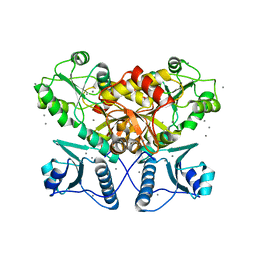 | |
5CRE
 
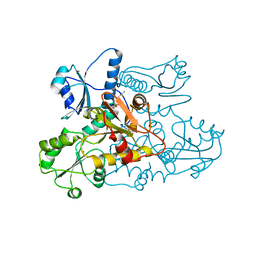 | | Human skeletal calsequestrin, D210G mutant low-calcium complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5CRD
 
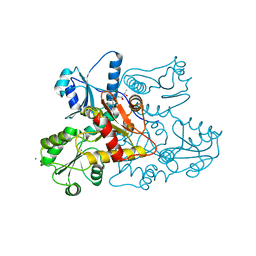 | | Wild-type human skeletal calsequestrin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
7SUX
 
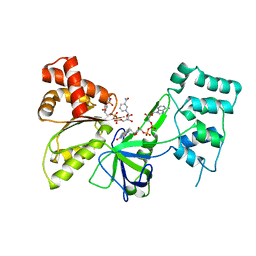 | |
7SV0
 
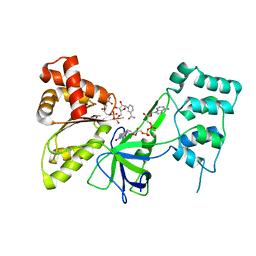 | |
7SUZ
 
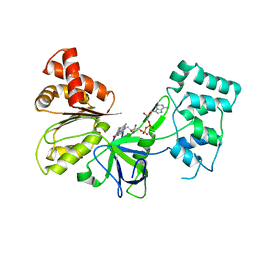 | |
6N14
 
 | | Phosphoserine BlaC, Class A serine beta-lactamase from Mycobacterium tuberculosis | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Moural, T.W, White, D.S, Choy, C.J, Kang, C, Berkman, C.E. | | Deposit date: | 2018-11-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.52169466 Å) | | Cite: | Crystal Structure of Phosphoserine BlaC fromMycobacterium tuberculosisInactivated by Bis(Benzoyl) Phosphate.
Int J Mol Sci, 20, 2019
|
|
5KVA
 
 | | Crystal Structure of sorghum caffeoyl-CoA O-methyltransferase (CCoAOMT) | | Descriptor: | CALCIUM ION, S-ADENOSYLMETHIONINE, caffeoyl-CoA O-methyltransferase | | Authors: | Walker, A.M, Sattler, S.A, Regner, M, Jones, J.P, Ralph, J, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Structure and Catalytic Mechanism of Sorghum bicolor Caffeoyl-CoA O-Methyltransferase.
Plant Physiol., 172, 2016
|
|
1C09
 
 | | RUBREDOXIN V44A CP | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Beard, B, Kang, C. | | Deposit date: | 1999-07-15 | | Release date: | 2001-02-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of the redox potential of the [Fe(SCys)(4)] site in rubredoxin by the orientation of a peptide dipole.
Biochemistry, 38, 1999
|
|
