1ZZE
 
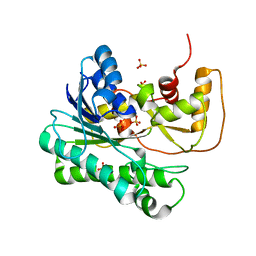 | | X-ray Structure of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor | | Descriptor: | Aldehyde reductase II, SULFATE ION | | Authors: | Kamitori, S, Iguchi, A, Ohtaki, A, Yamada, M, Kita, K. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
8WL7
 
 | |
8WL9
 
 | |
8WL5
 
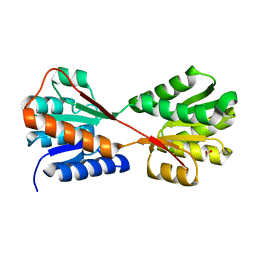 | | X-ray structure of Enterobacter cloacae allose-binding protein in free form | | Descriptor: | 1,2-ETHANEDIOL, Allose ABC transporter | | Authors: | Kamitori, S. | | Deposit date: | 2023-09-29 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structures of Enterobacter cloacae allose-binding protein in complexes with monosaccharides demonstrate its unique recognition mechanism for high affinity to allose.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8WLB
 
 | |
1JI2
 
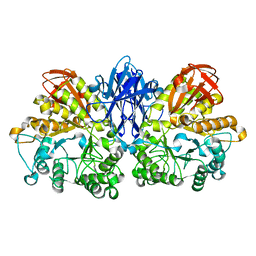 | | Improved X-ray Structure of Thermoactinomyces vulgaris R-47 alpha-Amylase 2 | | Descriptor: | ALPHA-AMYLASE II, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
1BVZ
 
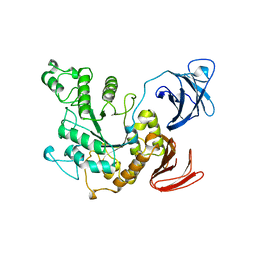 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
2GS9
 
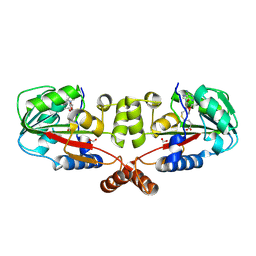 | | Crystal structure of TT1324 from Thermus thermophilis HB8 | | Descriptor: | FORMIC ACID, Hypothetical protein TT1324, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kamitori, S, Abe, A, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of TT1324 from Thermus thermophilis HB8
To be Published
|
|
1JI1
 
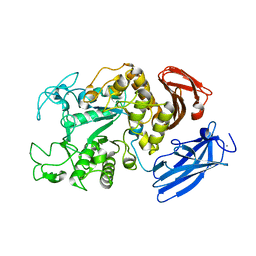 | | Crystal Structure Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 | | Descriptor: | ALPHA-AMYLASE I, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
173D
 
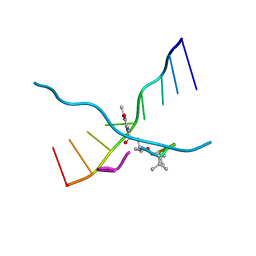 | | MULTIPLE BINDING MODES OF ANTICANCER DRUG ACTINOMYCIN D: X-RAY, MOLECULAR MODELING, AND SPECTROSCOPIC STUDIES OF D(GAAGCTTC)2-ACTINOMYCIN D COMPLEXES AND ITS HOST DNA | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Kamitori, S, Takusagawa, F. | | Deposit date: | 1994-04-18 | | Release date: | 1994-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple Binding Modes of Anticancer Drug Actinomycin D: X-Ray, Molecular Modeling, and Spectroscopic Studies of D(Gaagcttc)2-Actinomycin D Complexes and its Host DNA
J.Am.Chem.Soc., 116, 1994
|
|
172D
 
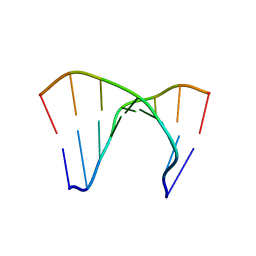 | |
2EIS
 
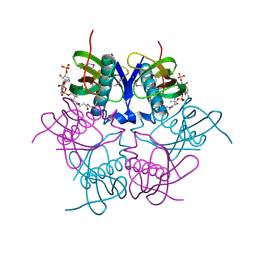 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
1IZE
 
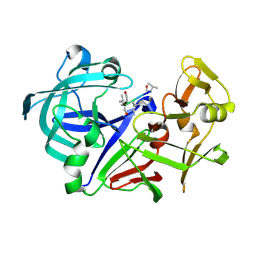 | | Crystal structure of Aspergillus oryzae Aspartic proteinase complexed with pepstatin | | Descriptor: | Pepstatin, alpha-D-mannopyranose, aspartic proteinase | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Aspergillus oryzae aspartic proteinase and its complex with an inhibitor pepstatin at 1.9A resolution.
J.Mol.Biol., 326, 2003
|
|
1IZD
 
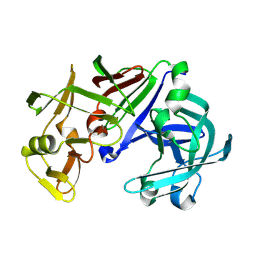 | | Crystal structure of Aspergillus oryzae Aspartic Proteinase | | Descriptor: | Aspartic proteinase, alpha-D-mannopyranose | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Aspergillus oryzae Aspartic Proteinase and its Complex with an Inhibitor Pepstatin at 1.9 A Resolution
J.Mol.Biol., 326, 2003
|
|
1UJM
 
 | |
1Y1P
 
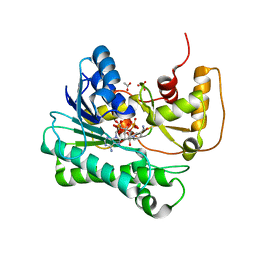 | | X-ray structure of aldehyde reductase with NADPH | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Aldehyde reductase II, ... | | Authors: | Kamitori, S, Kita, K. | | Deposit date: | 2004-11-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
8YXK
 
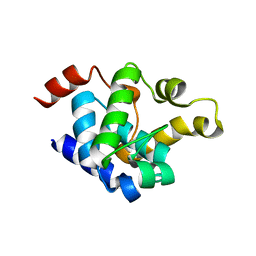 | |
8YXN
 
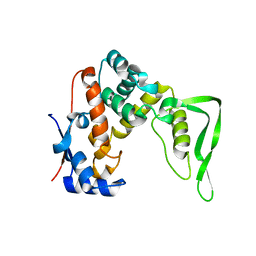 | |
8GSY
 
 | |
8GSX
 
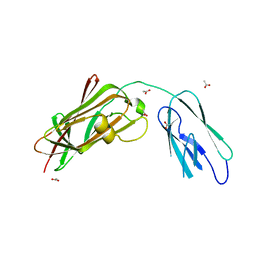 | |
7WRR
 
 | |
7WRT
 
 | |
6L67
 
 | | X-ray structure of human galectin-10 in complex with D-galactose | | Descriptor: | Galectin-10, beta-D-galactopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6B
 
 | | X-ray structure of human galectin-10 in complex with L-fucose | | Descriptor: | Galectin-10, beta-L-fucopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6C
 
 | | X-ray structure of human galectin-10 in complex with D-arabinose | | Descriptor: | Galectin-10, alpha-D-arabinopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
