7YZV
 
 | |
6QN1
 
 | | T=4 quasi-symmetric bacterial microcompartment particle | | Descriptor: | BMC domain-containing protein, Carbon dioxide concentrating mechanism protein CcmL | | Authors: | Kalnins, G. | | Deposit date: | 2019-02-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles.
Nat Commun, 11, 2020
|
|
9F3W
 
 | |
9F3Y
 
 | |
9F3X
 
 | |
6FF0
 
 | |
6FEZ
 
 | | Ryegrass mottle virus protease domain | | Descriptor: | Serine protease domain | | Authors: | Kalnins, G. | | Deposit date: | 2018-01-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | VPg Impact on Ryegrass Mottle Virus Serine-like 3C Protease Proteolysis and Structure.
Int J Mol Sci, 24, 2023
|
|
5A0Z
 
 | |
5A0U
 
 | |
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZD
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RVB
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV8
 
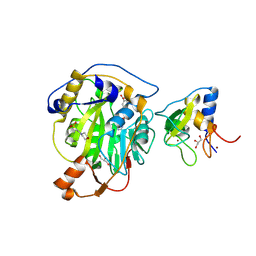 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 5 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV4
 
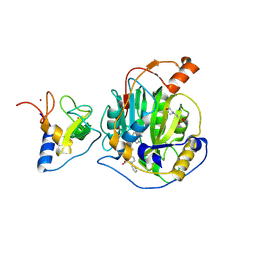 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV9
 
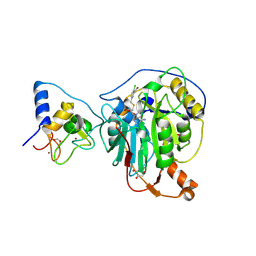 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 6 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-chloranyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV5
 
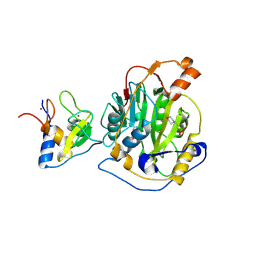 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 1 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV7
 
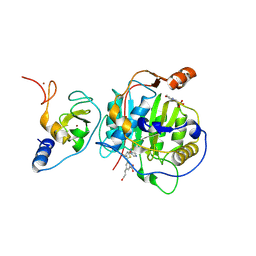 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 4 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-oxidanylprop-1-ynyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RV6
 
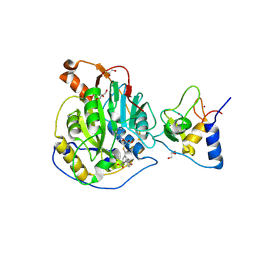 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(4-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RVA
 
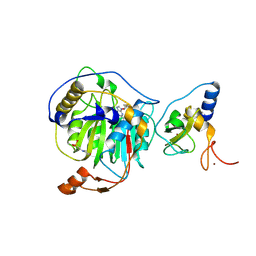 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 7 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6XUO
 
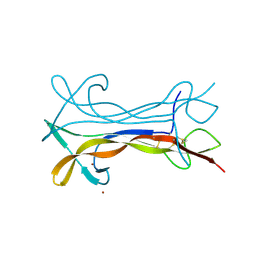 | | mature recombinant horse NGF | | Descriptor: | Nerve growth factor, ZINC ION | | Authors: | Kalnins, G. | | Deposit date: | 2020-01-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Mature horse nerve growth factor at 2.8 angstrom resolution
To Be Published
|
|
6YFB
 
 | | Virus-like particle of bacteriophage AVE016 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFI
 
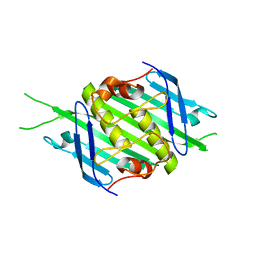 | | Bacteriophage EMS014 coat protein | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFJ
 
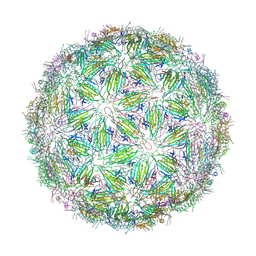 | | Virus-like particle of bacteriophage ESE001 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.233 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFP
 
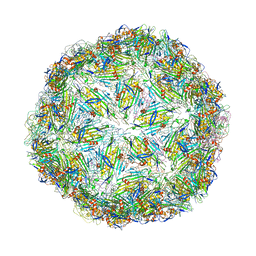 | | Virus-like particle of bacteriophage GQ-112 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
