5XXA
 
 | | beta-1,4-mannanase-SeMet-RmMan134A | | Descriptor: | endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, D, Huang, P. | | Deposit date: | 2017-07-03 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of endo-1,4-beta-mannanase at 1.76 Angstroms resolution.
To Be Published
|
|
5XWF
 
 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (SP3221/SAD) | | Descriptor: | Fungal chitinase from Rhizomucor miehei (SeMet-substituted proteins) | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
5YXT
 
 | |
7XMJ
 
 | |
7VTM
 
 | |
7VTL
 
 | |
7VTK
 
 | |
7WW4
 
 | |
7WWC
 
 | | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose | | Descriptor: | beta-1,3(4)-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2021625 Å) | | Cite: | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose
To Be Published
|
|
5XTJ
 
 | | Mannanase(RmMan134A) | | Descriptor: | Endo beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Huang, P. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
4K35
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5YLH
 
 | | Structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5XTT
 
 | | Crystal structure of RmMan134A-M3 complex | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XUL
 
 | | Complex structure (RmMan134A-M6). | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XUG
 
 | | Complex structure(RmMan134A-M5). | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XU5
 
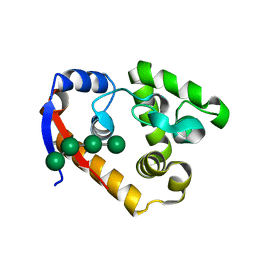 | | Complex structure of RmMan134A-M4 | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
3VM7
 
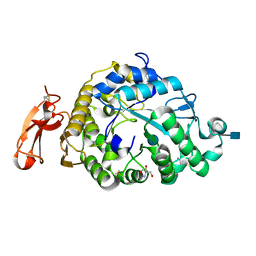 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
6JHI
 
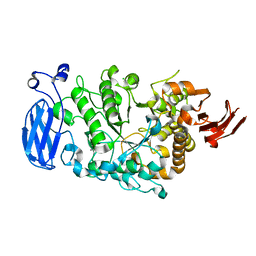 | | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose
To Be Published
|
|
6JHH
 
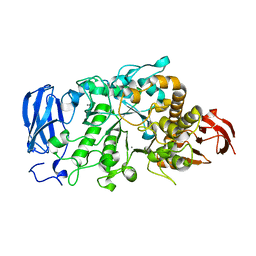 | | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose | | Descriptor: | CALCIUM ION, Pulullanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose
To Be Published
|
|
6JHG
 
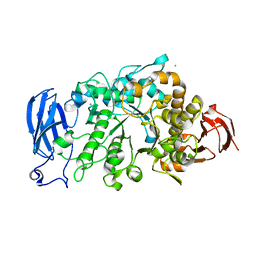 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
7Y1S
 
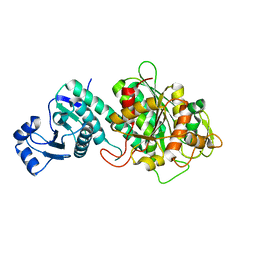 | |
7Y1X
 
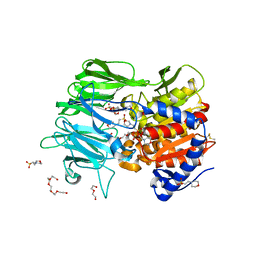 | |
4LYR
 
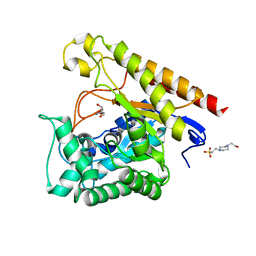 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
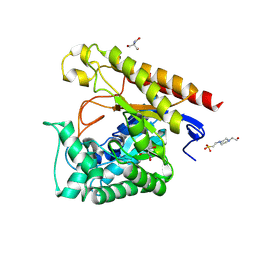 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
