2PJI
 
 | |
2A3S
 
 | |
1MR6
 
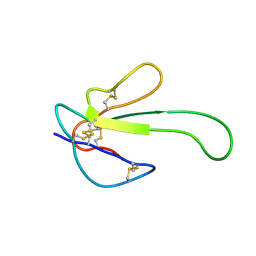 | | Solution Structure of gamma-Bungarotoxin:Implication for the role of the Residues Adjacent to RGD in Integrin Binding | | Descriptor: | neurotoxin | | Authors: | Chuang, W.-J, Shiu, J.-H, Chen, C.-Y, Chen, Y.-C, Chang, L.-S. | | Deposit date: | 2002-09-18 | | Release date: | 2004-05-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma-bungarotoxin: The functional significance of amino acid residues flanking the RGD motif in integrin binding
Proteins, 57, 2004
|
|
2D2W
 
 | |
2C6Y
 
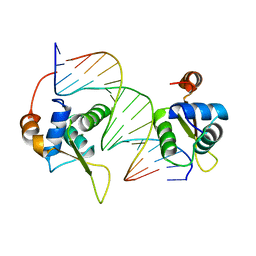 | | Crystal structure of interleukin enhancer-binding factor 1 bound to DNA | | Descriptor: | FORKHEAD BOX PROTEIN K2, INTERLEUKIN 2 PROMOTOR, MAGNESIUM ION | | Authors: | Tsai, K.-L, Huang, C.-Y, Chang, C.-H, Sun, Y.-J, Chuang, W.-J, Hsiao, C.-D. | | Deposit date: | 2005-11-15 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Foxk1A-DNA Complex and its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins.
J.Biol.Chem., 281, 2006
|
|
1XNZ
 
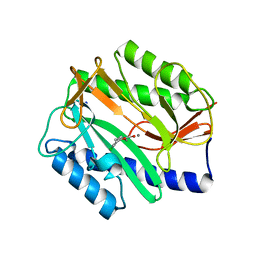 | | Crystal Structure of Mn(II) form of E. coli. Methionine Aminopeptidase in complex with 5-(2-chlorophenyl)furan-2-carboxylic acid | | Descriptor: | 5-(2-CHLOROPHENYL)FURAN-2-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z, Xie, S.-X, Huang, M, Huang, W.-J, Lu, J.-P, Ma, Z.-Q. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Metalloform-Selective Inhibitors of Escherichia coli Methionine Aminopeptidase and X-ray Structure of a Mn(II)-Form Enzyme Complexed with an Inhibitor.
J.Am.Chem.Soc., 126, 2004
|
|
1J3S
 
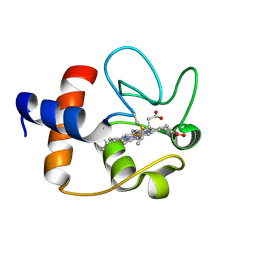 | |
2EVC
 
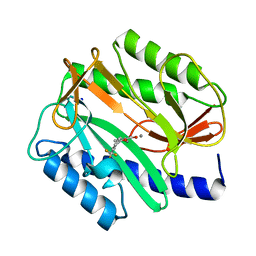 | |
2EVM
 
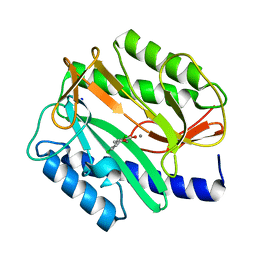 | |
2EVO
 
 | |
2ATH
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist | | Descriptor: | 2-{5-[3-(7-PROPYL-3-TRIFLUOROMETHYLBENZO[D]ISOXAZOL-6-YLOXY)PROPOXY]INDOL-1-YL}ETHANOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Mahindroo, N, Huang, C.-F, Wu, S.-Y, Hsieh, H.-P. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Novel indole-based peroxisome proliferator-activated receptor agonists: design, SAR, structural biology, and biological activities
J.Med.Chem., 48, 2005
|
|
4XH6
 
 | | Crystal structure of proto-oncogene kinase Pim1 bound to hispidulin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one, Serine/threonine-protein kinase pim-1 | | Authors: | Su, M.-Y, Chang, C.-I. | | Deposit date: | 2015-01-05 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Total Synthesis of Hispidulin and the Structural Basis for Its Inhibition of Proto-oncogene Kinase Pim-1
J.Nat.Prod., 78, 2015
|
|
