1AWW
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, 42 STRUCTURES | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
1AWX
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
5O2X
 
 | | Extended catalytic domain of H. jecorina LPMO9A a.k.a EG4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Glycoside hydrolase family 61, ... | | Authors: | Hansson, H, Karkehabadi, S, Mikkelsen, N.E, Sandgren, M, Kelemen, B, Kaper, T. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain.
J. Biol. Chem., 292, 2017
|
|
6GZY
 
 | | HOIP-fragment5 complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF31, SODIUM ION, ... | | Authors: | Johansson, H, Tsai, Y.C.I, Fantom, K, Chung, C.W, Martino, L, House, D, Rittinger, K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP.
J. Am. Chem. Soc., 141, 2019
|
|
5O2W
 
 | | Extended catalytic domain of Hypocrea jecorina LPMO 9A. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Glycoside hydrolase family 61, ... | | Authors: | Karkehabadi, S, Hansson, H, Sandgren, M, Mikelssen, N.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain.
J. Biol. Chem., 292, 2017
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
3ZZ1
 
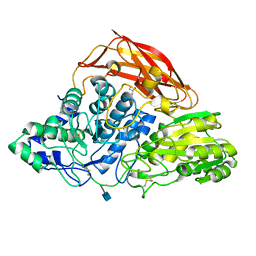 | | Crystal structure of a glycoside hydrolase family 3 beta-glucosidase, Bgl1 from Hypocrea jecorina at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCOSIDE GLUCOHYDROLASE, GLYCEROL | | Authors: | Sandgren, M, Kaper, T, Mikkelsen, N.E, Hansson, H, Piens, K, Gudmundsson, M, Larenas, E, Kelemen, B, Karkehabadi, S. | | Deposit date: | 2011-08-31 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 Beta-Glucosidase, Cel3A from Hypocrea Jecorina.
J.Biol.Chem., 289, 2014
|
|
3ZYZ
 
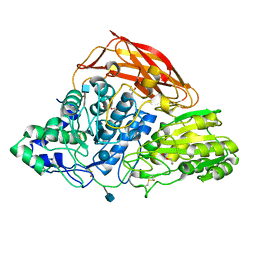 | | Crystal structure of a glycoside hydrolase family 3 beta-glucosidase, Bgl1 from Hypocrea jecorina at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCOSIDE GLUCOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandgren, M, Kaper, T, Mikkelsen, N.E, Hansson, H, Piens, K, Gudmundsson, M, Larenas, E, Kelemen, B, Karkehabadi, S. | | Deposit date: | 2011-08-30 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 Beta-Glucosidase, Cel3A from Hypocrea Jecorina.
J.Biol.Chem., 289, 2014
|
|
8POF
 
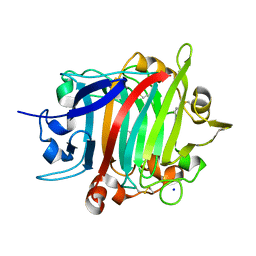 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | Authors: | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
2YG1
 
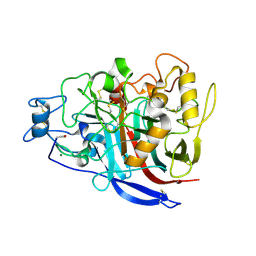 | | APO STRUCTURE OF CELLOBIOHYDROLASE 1 (CEL7A) FROM HETEROBASIDION ANNOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, MAGNESIUM ION | | Authors: | Haddad-Momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
2XSP
 
 | | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, ... | | Authors: | Haddad-momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
2VN7
 
 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
2VTC
 
 | | The structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEL61B, NICKEL (II) ION | | Authors: | Karkehabadi, S, Hansson, H, Kim, S, Piens, K, Mitchinson, C, Sandgren, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The First Structure of a Glycoside Hydrolase Family 61 Member, Cel61B from the Hypocrea Jecorina, at 1.6 A Resolution.
J.Mol.Biol., 383, 2008
|
|
5AE6
 
 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) in complex with 4-thioxylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-XYLOSIDASE, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
2VN4
 
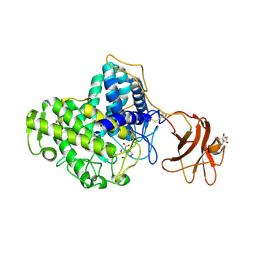 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
5A7M
 
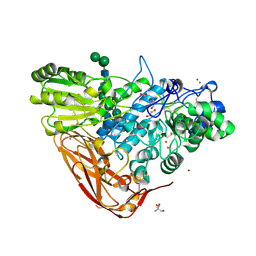 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
5O5D
 
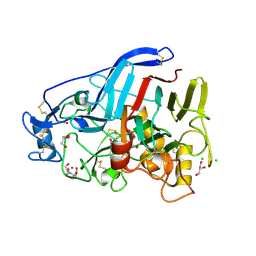 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
2WNE
 
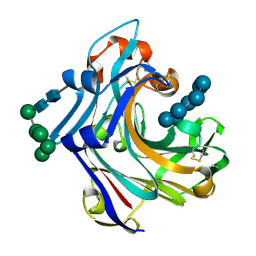 | | Mutant Laminarinase 16A cyclizes laminariheptaose | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H.M, Hansson, H, Engstrom, A, Einarsson, E, Forsberg, Z, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-07-09 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
5O59
 
 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 1-thio-beta-D-glucopyranose, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
5OA5
 
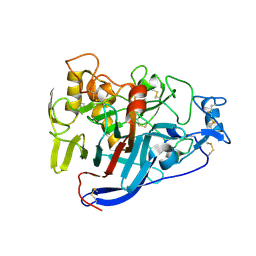 | | CELLOBIOHYDROLASE I (CEL7A) FROM HYPOCREA JECORINA WITH IMPROVED THERMAL STABILITY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, GLYCEROL | | Authors: | Goedegebuur, F, Hansson, H, Karkehabadi, S, Mikkelsen, N, Stahlberg, J, Sandgren, M. | | Deposit date: | 2017-06-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution.
J. Biol. Chem., 292, 2017
|
|
2WLQ
 
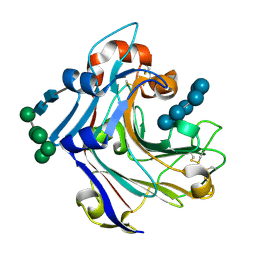 | | Nucleophile-disabled Lam16A mutant holds laminariheptaose (L7) in a cyclical conformation | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H, Hansson, H, Engstrom, A, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
4CSI
 
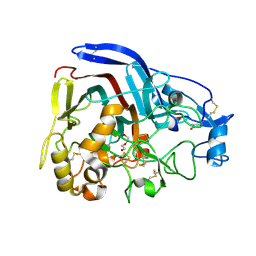 | | Crystal structure of the thermostable Cellobiohydrolase Cel7A from the fungus Humicola grisea var. thermoidea. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULASE, DI(HYDROXYETHYL)ETHER | | Authors: | Haddad-Momeni, M, Goedegebuur, F, Hansson, H, Karkehabadi, S, Askarieh, G, Mitchinson, C, Larenas, E, Stahlberg, J, Sandgren, M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Crystal Structure and Cellulase Activity of the Thermostable Cellobiohydrolase Cel7A from the Fungus Humicola Grisea Var. Thermoidea.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AX6
 
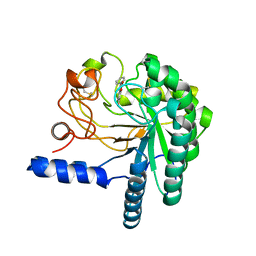 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AX7
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AU0
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
