7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
6K8I
 
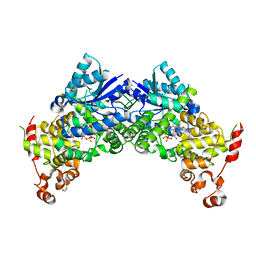 | | Crystal structure of Arabidopsis thaliana CRY2 | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, L, Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5IL1
 
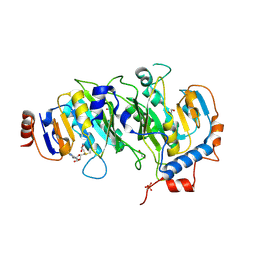 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
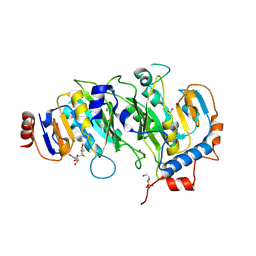 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
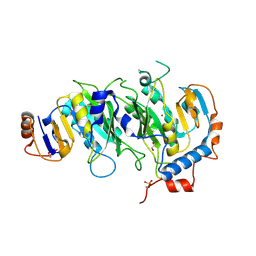 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
6K8K
 
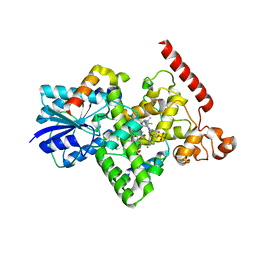 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5I9D
 
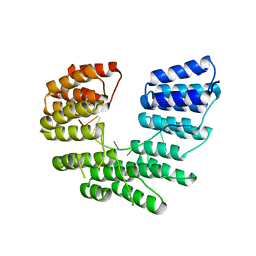 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9F
 
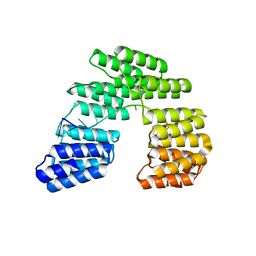 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9H
 
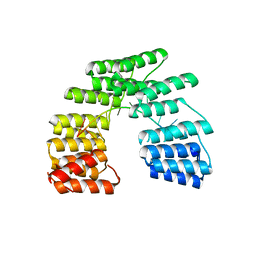 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
4J72
 
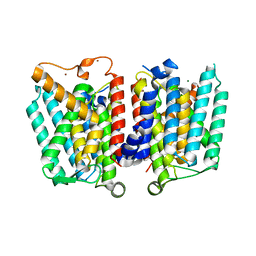 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
5GP0
 
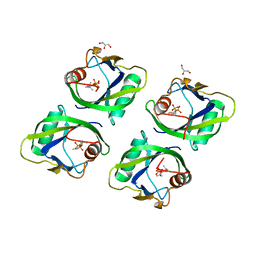 | | Crystal structure of geraniol-NUDX1 complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, Nudix hydrolase 1 | | Authors: | Liu, J, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-07-30 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5GI0
 
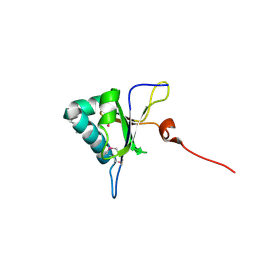 | | Crystal structure of RNA editing factor MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5ZT3
 
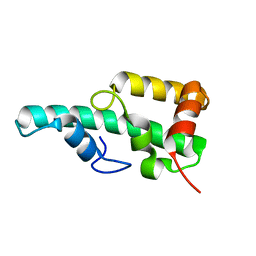 | | Crystal structure of WA352 from Oryza sativa | | Descriptor: | WA352 | | Authors: | Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice
Biochem. Biophys. Res. Commun., 501, 2018
|
|
5GHE
 
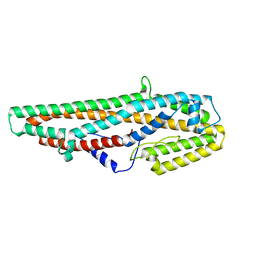 | | Crystal Structure of Bacillus thuringiensis Cry6Aa2 Protoxin | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Huang, J, Guan, Z, Zou, T, Sun, M, Yin, P. | | Deposit date: | 2016-06-19 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of Cry6Aa: A novel nematicidal ClyA-type alpha-pore-forming toxin from Bacillus thuringiensis
Biochem.Biophys.Res.Commun., 478, 2016
|
|
5ISY
 
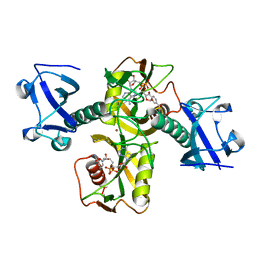 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
5IWW
 
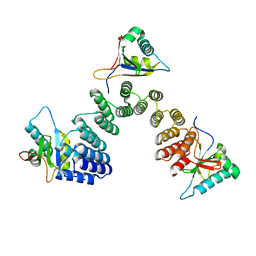 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5IWB
 
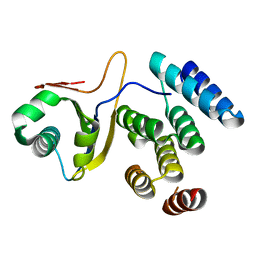 | | Crystal structure of design pentatricopeptide repeat complex with MORF protein | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS3-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis for designer pentatricopeptide repeat proteins interaction with MORF protein
To be published
|
|
5IZW
 
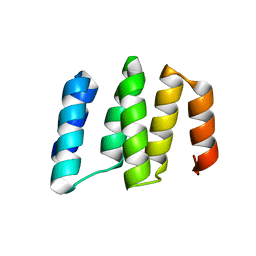 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5WWD
 
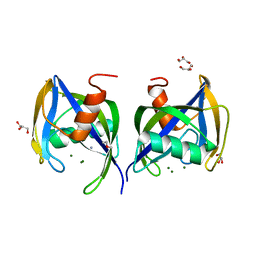 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
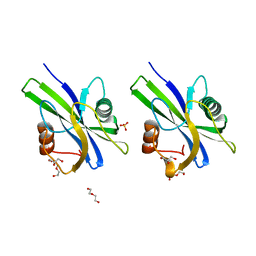 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
7C9O
 
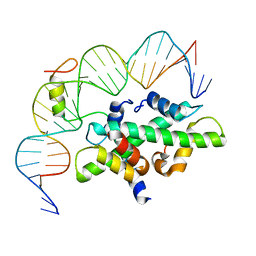 | | Crystal structure of DNA-bound CCT/NF-YB/YC complex (HD1CCT/GHD8/OsNF-YC2) | | Descriptor: | DNA (25-MER), Nuclear transcription factor Y subunit B-11, Nuclear transcription factor Y subunit C-2, ... | | Authors: | Shen, C, Liu, H, Guan, Z, Xing, Y, Yin, P. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering.
Plant Cell, 32, 2020
|
|
7C9P
 
 | | Crystal structure of rice histone-fold dimer GHD8/OsNF-YC2 | | Descriptor: | Nuclear transcription factor Y subunit B-11, Nuclear transcription factor Y subunit C-2 | | Authors: | Shen, C, Liu, H, Guan, Z, Xing, Y, Yin, P. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering.
Plant Cell, 32, 2020
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
