2KSY
 
 | | Solution nmr structure of sensory rhodopsin II | | Descriptor: | RETINAL, Sensory rhodopsin II | | Authors: | Gautier, A, Mott, H.R, Bostock, M.J, Kirkpatrick, J.P, Nietlispach, D. | | Deposit date: | 2010-01-14 | | Release date: | 2010-06-02 | | Last modified: | 2015-06-10 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4BAD
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxymethyltriazoledipicolinate complex at 1.35 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BAR
 
 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxyethyltriazoledipicolinate complex at 1.20 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Clicked europium dipicolinate complexes for protein X-ray structure determination.
Chem. Commun. (Camb.), 48, 2012
|
|
4BAF
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethyltriazoledipicolinate complex at 1.51 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BAP
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethylcholinetriazoledipicolinate complex at 1.21 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BAL
 
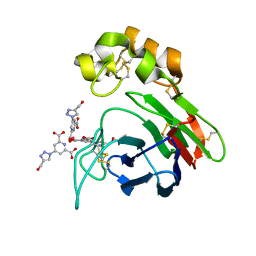 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxymethyltriazoledipicolinate complex at 1.30 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4WCT
 
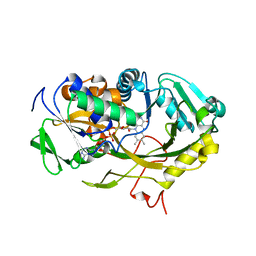 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
4XWZ
 
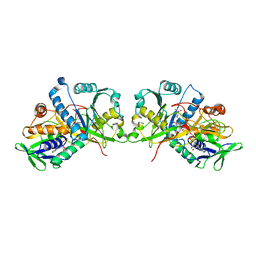 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus in complex with the substrate fructosyl lysine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase, LYSINE, ... | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2015-01-29 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
6Y4J
 
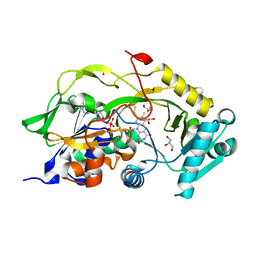 | | Engineered Fructosyl Peptide Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase, GLYCEROL, ... | | Authors: | Donini, S, Rigoldi, F, Gautieri, A, Parisini, E. | | Deposit date: | 2020-02-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Rational backbone redesign of a fructosyl peptide oxidase to widen its active site access tunnel.
Biotechnol.Bioeng., 117, 2020
|
|
5OC2
 
 | | Crystal structure of Asp295Cys/Lys303Cys Amadoriase I mutant from Aspergillus Fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Donini, S, Gautieri, A, Parisini, E. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Thermal stabilization of the deglycating enzyme Amadoriase I by rational design.
Sci Rep, 8, 2018
|
|
5OC3
 
 | | Crystal structure of Ser67Cys/Pro121Cys Amadoriase I mutant from Aspergillus Fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase, GLYCEROL | | Authors: | Rigoldi, F, Donini, S, Gautieri, A, Parisini, E. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Thermal stabilization of the deglycating enzyme Amadoriase I by rational design.
Sci Rep, 8, 2018
|
|
8BJY
 
 | | Engineered Fructosyl Peptide Oxidase - X02B mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02B), GLYCEROL | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BLX
 
 | | Engineered Fructosyl Peptide Oxidase - X02A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02A), ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BMU
 
 | | Engineered Fructosyl Peptide Oxidase - X04 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X04), ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BLZ
 
 | | Engineered Fructosyl Peptide Oxidase - D02 mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (D02), GLYCEROL, ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
