1DFA
 
 | | CRYSTAL STRUCTURE OF PI-SCEI IN C2 SPACE GROUP | | Descriptor: | PI-SCEI ENDONUCLEASE | | Authors: | Hu, D, Crist, M, Duan, X, Quiocho, F.A, Gimble, F.S. | | Deposit date: | 1999-11-18 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the structure of the PI-SceI-DNA complex by affinity cleavage and affinity photocross-linking.
J.Biol.Chem., 275, 2000
|
|
6B7U
 
 | | Structure of hen egg-white lysozyme without high-pressure pre-treatment | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
6B7V
 
 | | Structure of hen egg-white lysozyme pre-treated with high-pressure homogenization at 120 MPa | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
6B7W
 
 | | Structure of hen egg-white lysozyme pre-treated with high pressure (600 MPa) under isobaric condition | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
5AKC
 
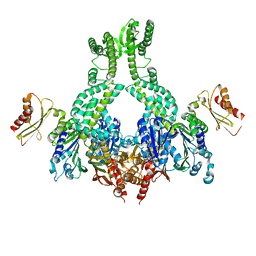 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKD
 
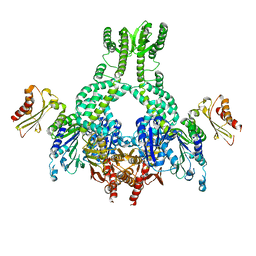 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKB
 
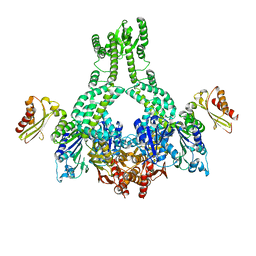 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
8BN3
 
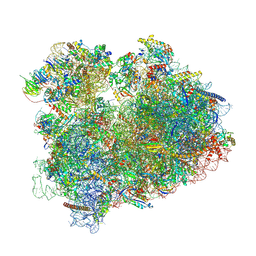 | | Yeast 80S, ES7s delta, eIF5A, Stm1 containing | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Dimitrova-Paternoga, L, Paternoga, H, Wilson, D.N. | | Deposit date: | 2022-11-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Evolving precision: rRNA expansion segment 7S modulates translation velocity and accuracy in eukaryal ribosomes.
Nucleic Acids Res., 52, 2024
|
|
5AIM
 
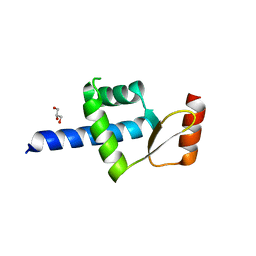 | | Crystal structure of T138 central eWH domain | | Descriptor: | GLYCEROL, TRANSCRIPTION FACTOR TAU 138 KDA SUBUNIT | | Authors: | Male, G, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly.
Nat Commun, 6, 2015
|
|
5AIO
 
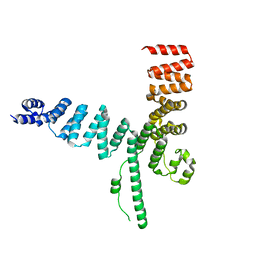 | |
5AEM
 
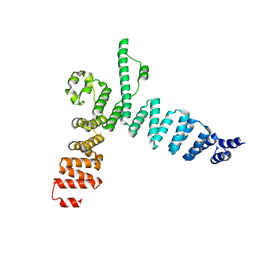 | | Structure of t131 N-terminal TPR array | | Descriptor: | TRANSCRIPTION FACTOR TAU 131 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Muller, C.W. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly.
Nat Commun, 6, 2015
|
|
