4W9R
 
 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
5JH8
 
 | | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472 | | Descriptor: | (2S)-2-(dimethylamino)-4-(methylselanyl)butanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chang, C, Michalska, K, Tesar, C, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.018 Å) | | Cite: | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472
To Be Published
|
|
4LZK
 
 | |
4M7O
 
 | |
3NBM
 
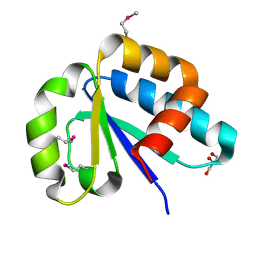 | | The lactose-specific IIB component domain structure of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) from Streptococcus pneumoniae. | | Descriptor: | GLYCEROL, PTS system, lactose-specific IIBC components | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the lactose-specific IIB component domain of the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) from Streptococcus pneumoniae.
TO BE PUBLISHED
|
|
3O5V
 
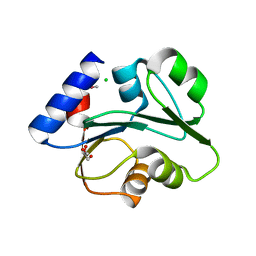 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
5BMQ
 
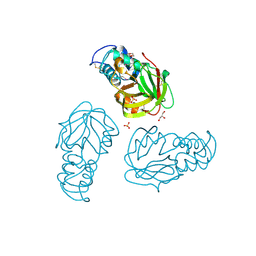 | | Crystal structure of L,D-transpeptidase (Yku) from Stackebrandtia nassauensis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ErfK/YbiS/YcfS/YnhG family protein, GLYCEROL, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of L,D-transpeptidases (Yku)
To be Published
|
|
5C4Y
 
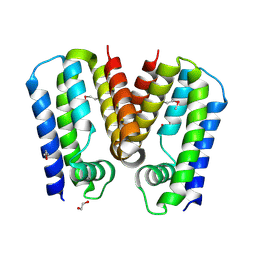 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
2ZC2
 
 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
4W66
 
 | |
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | Authors: | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
6Q2B
 
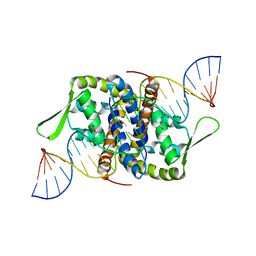 | | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA | | Descriptor: | ACETIC ACID, DNA (26-MER), MarR family transcriptional regulator | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA.
To Be Published
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
5JMU
 
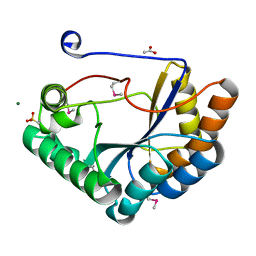 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
2OTD
 
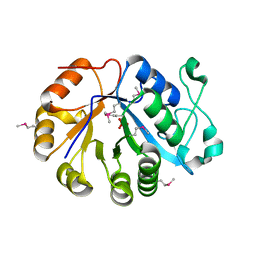 | | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a | | Descriptor: | Glycerophosphodiester phosphodiesterase, PHOSPHATE ION | | Authors: | Zhang, R, Wu, R, Clancy, S, Jiang, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a
To be Published
|
|
5CJ3
 
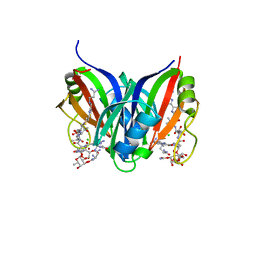 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
5DUK
 
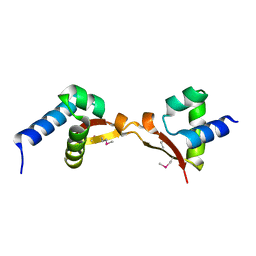 | |
5UQP
 
 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | Descriptor: | CHLORIDE ION, Cupin, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
2OQT
 
 | | Structural Genomics, the crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS | | Descriptor: | Hypothetical protein SPy0176 | | Authors: | Tan, K, Wu, R, Osipiuk, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS
To be Published
|
|
2OZZ
 
 | |
2OT9
 
 | |
2P0T
 
 | | Structural Genomics, the crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, UPF0307 protein PSPTO_4464 | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000
To be Published
|
|
5HX0
 
 | | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053 | | Descriptor: | ACETATE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Duke, N, Clancy, S, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053
To Be Published
|
|
5I47
 
 | | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, RimK domain protein ATP-grasp | | Authors: | Chang, C, Duke, N, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745
To Be Published
|
|
5IX8
 
 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
