6REI
 
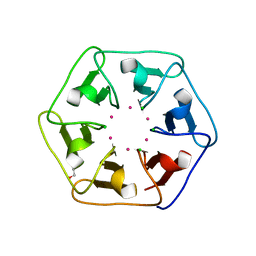 | | Crystal structure of Pizza6-S with Cd2+ | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REM
 
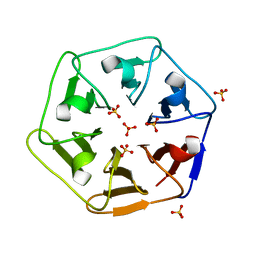 | | Crystal structure of Pizza6-SH with Sulphate ion | | Descriptor: | Pizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REL
 
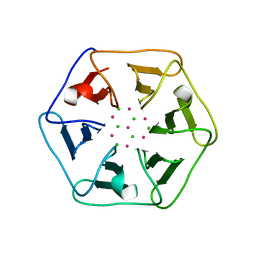 | | Crystal structure of Pizza6-SH with CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REO
 
 | | Crystal structure of 3fPizza6-SH with Sulphate ion | | Descriptor: | 3fPizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
2G87
 
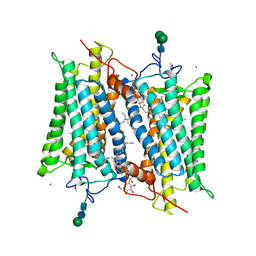 | | Crystallographic model of bathorhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic analysis of primary visual photochemistry
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2RT8
 
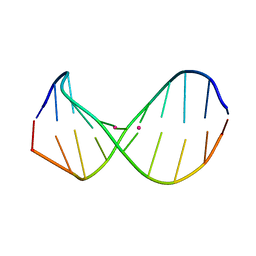 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
3VRE
 
 | | The crystal structure of hemoglobin from woolly mammoth in the deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VRG
 
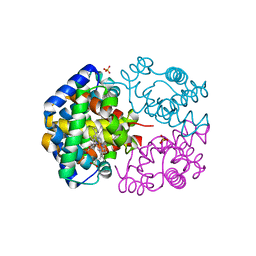 | | The crystal structure of hemoglobin from woolly mammoth in the met form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VRF
 
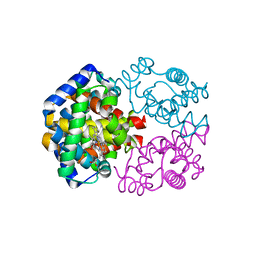 | | The crystal structure of hemoglobin from woolly mammoth in the carbonmonoxy forms | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
1GLV
 
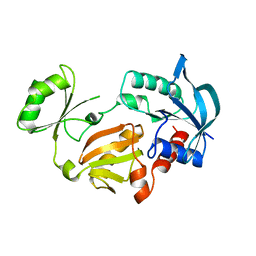 | |
1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
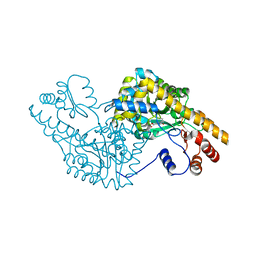
1G4V
 
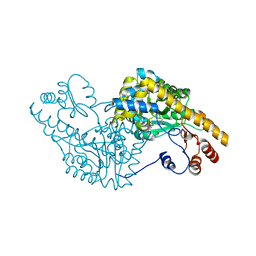 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G7W
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4X
 
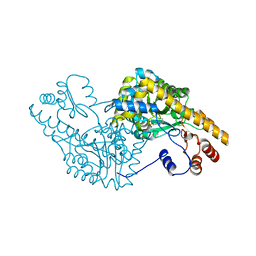 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
2H9V
 
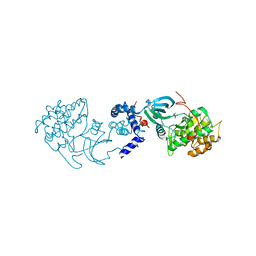 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1H8M
 
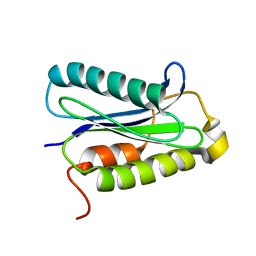 | |
1N2C
 
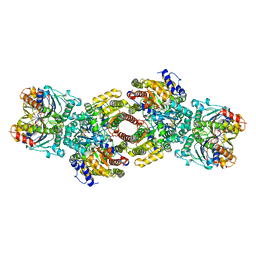 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schindelin, H, Kisker, C, Rees, D.C. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
1QLC
 
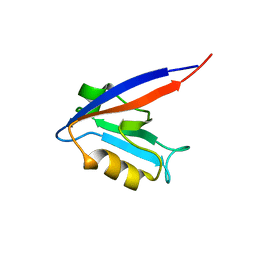 | |
2HPY
 
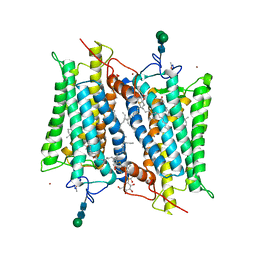 | | Crystallographic model of lumirhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local peptide movement in the photoreaction intermediate of rhodopsin
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JYY
 
 | |
6RLH
 
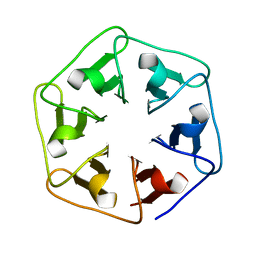 | |
6RLI
 
 | |
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
