4UV3
 
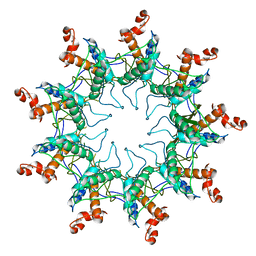 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4UV2
 
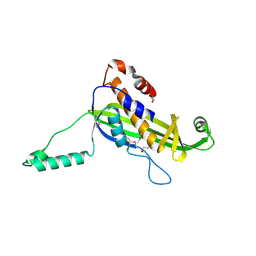 | | Structure of the curli transport lipoprotein CsgG in a non-lipidated, pre-pore conformation | | Descriptor: | CURLI PRODUCTION TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4XGC
 
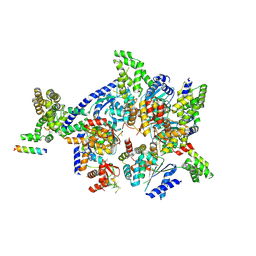 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
1RFY
 
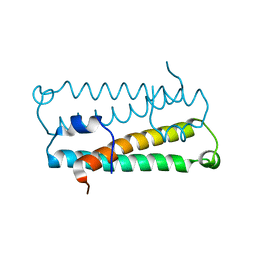 | | Crystal Structure of Quorum-Sensing Antiactivator TraM | | Descriptor: | Transcriptional repressor traM | | Authors: | Chen, G, Malenkos, J.W, Cha, M.R, Fuqua, C, Chen, L. | | Deposit date: | 2003-11-10 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Mol.Microbiol., 52, 2004
|
|
1AAF
 
 | | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Henderson, L.E, Chance, M.R, Bess Junior, J.W, South, T.L, Blake, P.R, Sagi, I, Perez-Alvarado, G, Sowder, R.C, Hare, D.R, Arthur, L.O. | | Deposit date: | 1992-04-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1.
Protein Sci., 1, 1992
|
|
3C8E
 
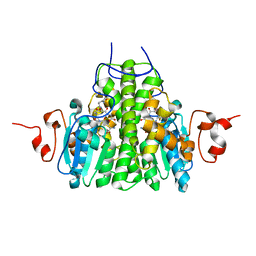 | | Crystal Structure Analysis of yghU from E. Coli | | Descriptor: | GLUTATHIONE, yghU, glutathione S-transferase homologue | | Authors: | Harp, J, Ladner, J.E, Schaab, M.R, Stourman, N.V, Armstrong, R.N. | | Deposit date: | 2008-02-11 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of YghU, a Nu-Class Glutathione Transferase Related to YfcG from Escherichia coli.
Biochemistry, 50, 2011
|
|
3K9A
 
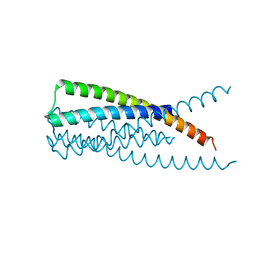 | | Crystal Structure of HIV gp41 with MPER | | Descriptor: | HIV glycoprotein gp41 | | Authors: | Shi, W, Han, D, Habte, H, Cho, M, Chance, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HIV gp41 with the membrane-proximal external region
J.Biol.Chem., 285, 2010
|
|
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
2WHW
 
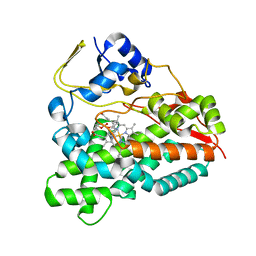 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLOTRIDECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WI9
 
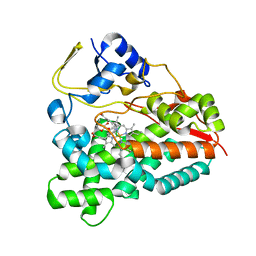 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLODODECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4NVQ
 
 | | Human G9a in Complex with Inhibitor A-366 | | Descriptor: | 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Sweis, R.F, Pliushchev, M, Brown, P.J, Guo, J, Li, F, Maag, D, Petros, A.M, Soni, N.B, Tse, C, Vedadi, M, Michaelides, M.R, Chiang, G.G, Pappano, W.N. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and development of potent and selective inhibitors of histone methyltransferase g9a.
ACS Med Chem Lett, 5, 2014
|
|
4F2D
 
 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
2XR7
 
 | | Crystal Structure of Nicotiana tabacum malonyltransferase (NtMat1) complexed with malonyl-coa | | Descriptor: | MALONYL-COENZYME A, MALONYLTRANSFERASE | | Authors: | Manjasetty, B.A, Yu, X.H, Panjikar, S, Taguchi, G, Chance, M.R, Liu, C.J. | | Deposit date: | 2010-09-11 | | Release date: | 2011-09-21 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Modification of Flavonol and Naphthol Glucoconjugates by Nicotiana Tabacum Malonyltransferase (Ntmat1).
Planta, 236, 2012
|
|
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1OMI
 
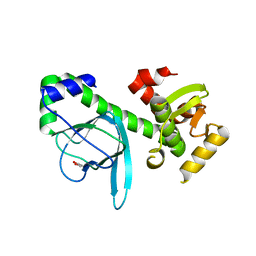 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
2AJT
 
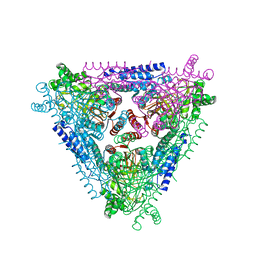 | | Crystal structure of L-Arabinose Isomerase from E.coli | | Descriptor: | L-arabinose isomerase | | Authors: | Manjasetty, B.A, Fedorov, E.V, Almo, S.C, Chance, M.R, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli L-Arabinose Isomerase (ECAI), The Putative Target of Biological Tagatose Production
J.Mol.Biol., 360, 2006
|
|
2NNU
 
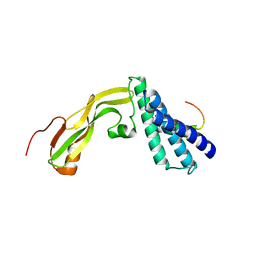 | |
1QQH
 
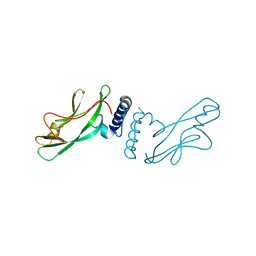 | |
1TUE
 
 | |
7T67
 
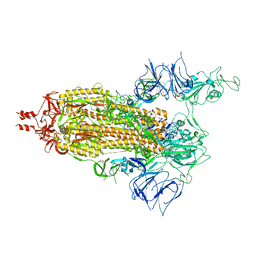 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7T3M
 
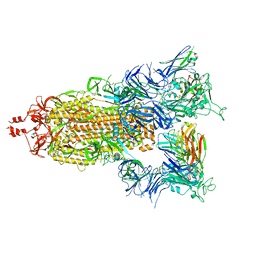 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2-7 scFv, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
1JR7
 
 | |
2P7O
 
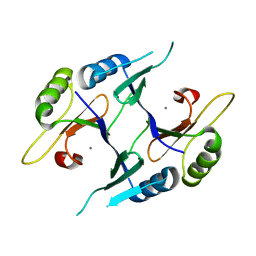 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes (tetragonal form) | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P4U
 
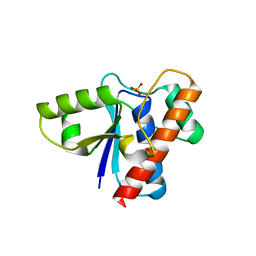 | | Crystal structure of acid phosphatase 1 (Acp1) from Mus musculus | | Descriptor: | Acid phosphatase 1, PHOSPHATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2PBN
 
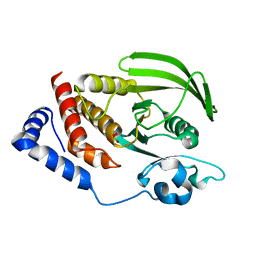 | | Crystal structure of the human tyrosine receptor phosphate gamma | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Reyes, C, Pelletier, L, Jin, X, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
