2YHJ
 
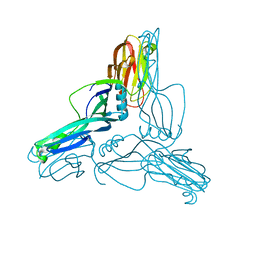 | | Clostridium perfringens Enterotoxin at 4.0 Angstrom Resolution | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, McClane, B.A, Basak, A.K. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
2XH6
 
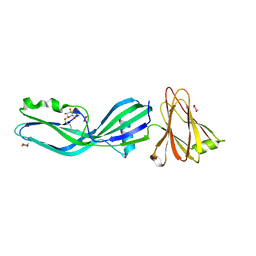 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
6FPY
 
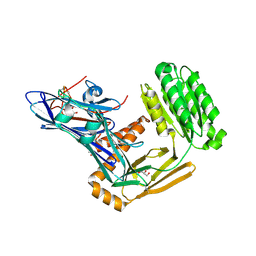 | | Inter-alpha-inhibitor heavy chain 1, wild type | | Descriptor: | GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1, MAGNESIUM ION | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
6FPZ
 
 | | Inter-alpha-inhibitor heavy chain 1, D298A | | Descriptor: | ACETATE ION, GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1 | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
5IK4
 
 | | Laminin A2LG45 C-form, Apo. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
5IK7
 
 | | Laminin A2LG45 I-form, Apo. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
5IK5
 
 | | Laminin A2LG45 C-form, G6/7 bound. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
5IK8
 
 | | Laminin A2LG45 I-form, G6/7 bound. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
2WNO
 
 | | X-ray Structure of CUB_C domain from TSG-6 | | Descriptor: | CALCIUM ION, COBALT (II) ION, TUMOR NECROSIS FACTOR-INDUCIBLE GENE 6 PROTEIN | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal Ion-Dependent Heavy Chain Transfer Activity of Tsg-6 Mediates Assembly of the Cumulus-Oocyte Matrix.
J.Biol.Chem., 290, 2015
|
|
6EJC
 
 | |
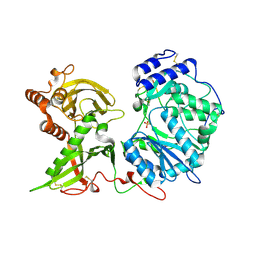
6EJE
 
 | |
6EJ8
 
 | |
6EJD
 
 | |
6EJB
 
 | |
6EJ7
 
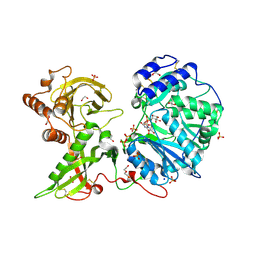 | |
6EJA
 
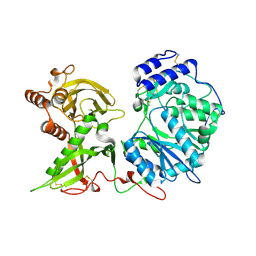 | |
6EJ9
 
 | |
6FOA
 
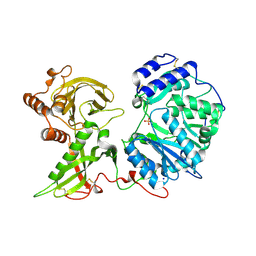 | | Human Xylosyltransferase 1 apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Structural Basis for the Initiation of Glycosaminoglycan Biosynthesis by Human Xylosyltransferase 1.
Structure, 26, 2018
|
|
7QSR
 
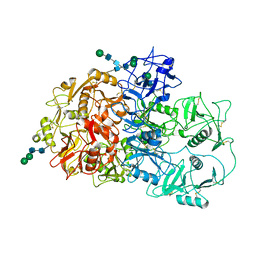 | | CryoEM structure of the Ectodomain of Human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Briggs, D.C, Lockhart-Cairns, M.P, Baldock, C. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NZN
 
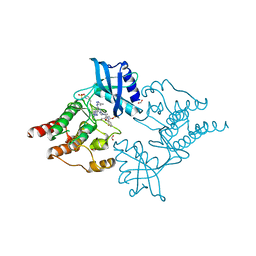 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
2J17
 
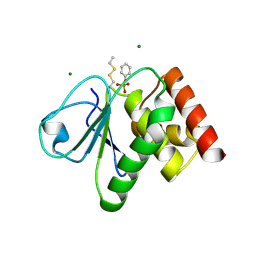 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2IYB
 
 | |
2J16
 
 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
1V1P
 
 | |
1OLP
 
 | | Alpha Toxin from Clostridium Absonum | | Descriptor: | ALPHA-TOXIN, CALCIUM ION, ZINC ION | | Authors: | Briggs, D.C, Basak, A.K. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clostridium Absonum Alpha-Toxin: New Insights Into Clostridial Phospholipase C Substrate Binding and Specificity
J.Mol.Biol., 333, 2003
|
|
