4DLE
 
 | | Ternary Structure of the large Fragment of Taq DNA Polymerase: 4-Fluoroproline Variant | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA polymerase I, ... | | Authors: | Holzberger, B, Obeid, S, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-02-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into the potential of 4-fluoroproline to modulate biophysical properties of protein
Chem Sci, 3, 2012
|
|
4DLG
 
 | | Ternary Structure of the large Fragment of Taq DNA polymerase | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ACETATE ION, DNA polymerase I, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S, Holzberger, B. | | Deposit date: | 2012-02-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the potential of 4-fluoroproline to modulate biophysical properties of protein
Chem Sci, 3, 2012
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTK
 
 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTS
 
 | | Stalk of radial spoke 1 attached with doublet microtubule from Chlamydomonas reinhardtii | | Descriptor: | Calmodulin, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5C37
 
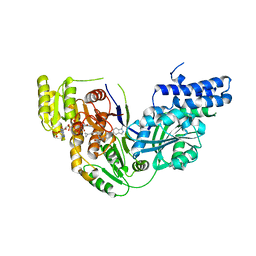 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | Descriptor: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | Authors: | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy.
Bioorg.Med.Chem.Lett., 28, 2018
|
|
5MUS
 
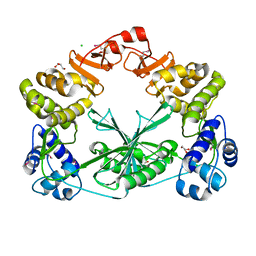 | | Structure of the C-terminal domain of a reptarenavirus L protein | | Descriptor: | CHLORIDE ION, GLYCEROL, L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
5MUZ
 
 | | Structure of a C-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
5MUY
 
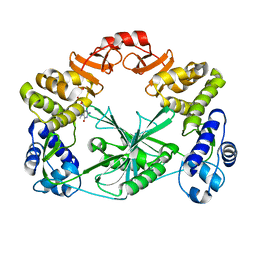 | | Structure of a C-terminal domain of a reptarenavirus L protein with m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
5MV0
 
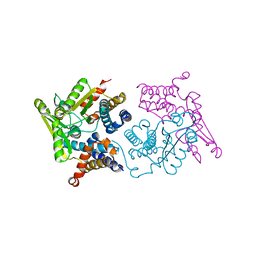 | | Structure of an N-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein, PHOSPHATE ION | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
6X9H
 
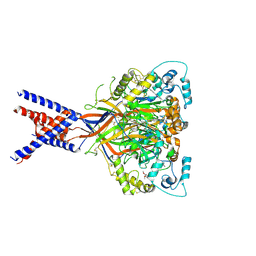 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
2LUJ
 
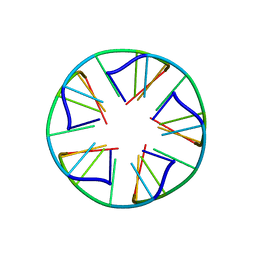 | | Solution structure of a parallel-stranded oligoisoguanine DNA pentaplex formed by d(T(iG)4T) in the presence of Cs ions | | Descriptor: | DNA (5'-D(*TP*(IGU)P*(IGU)P*(IGU)P*(IGU)P*T)-3') | | Authors: | Kang, M, Heuberger, B, Chaput, J.C, Switzer, C, Feigon, J. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Parallel-Stranded Oligoisoguanine DNA Pentaplex Formed by d(T(iG)4T) in the Presence of Cs(+) Ions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6E6B
 
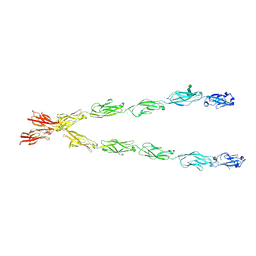 | | Crystal structure of the Protocadherin GammaB4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2018-07-24 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.52 Å) | | Cite: | Visualization of clustered protocadherin neuronal self-recognition complexes.
Nature, 569, 2019
|
|
5ZGG
 
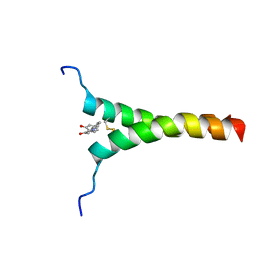 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
7AYI
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2a) | | Descriptor: | 7-(2-phenylazanylpyrimidin-4-yl)-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
7AYH
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
7P2P
 
 | | Human Signal Peptidase Complex Paralog A (SPC-A) | | Descriptor: | Signal peptidase complex catalytic subunit SEC11A, Signal peptidase complex subunit 1, Signal peptidase complex subunit 2, ... | | Authors: | Liaci, A.M, Foerster, F. | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage.
Mol.Cell, 81, 2021
|
|
7P2Q
 
 | | Human Signal Peptidase Complex Paralog C (SPC-C) | | Descriptor: | Signal peptidase complex catalytic subunit SEC11C, Signal peptidase complex subunit 1, Signal peptidase complex subunit 2, ... | | Authors: | Liaci, A.M, Foerster, F. | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage.
Mol.Cell, 81, 2021
|
|
7OGG
 
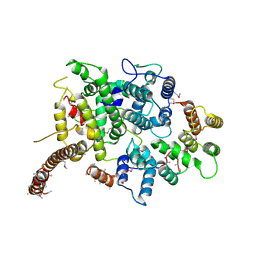 | | Nse5/6 complex | | Descriptor: | DNA repair protein KRE29,DNA repair protein KRE29,DNA repair protein KRE29, Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5 | | Authors: | Basquin, J, Taschner, M, Gruber, S. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding.
Embo J., 40, 2021
|
|
8Q7E
 
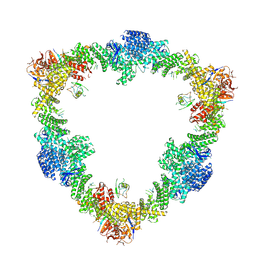 | |
8Q7H
 
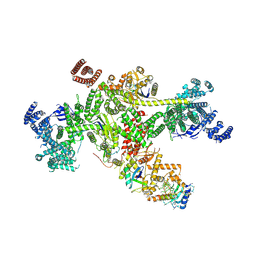 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4U38
 
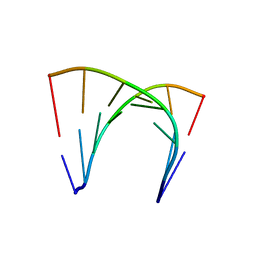 | | RNA duplex containing UU mispair | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4U34
 
 | | Crystal Structures of RNA Duplexes Containing 2-thio-Uridine | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*(SUR)P*CP*C-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4U35
 
 | | Crystal Structures of RNA Duplexes Containing 2-thio-Uridine | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*(SUR)P*CP*C-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4U37
 
 | | Native 7mer-RNA duplex | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
