1QOU
 
 | |
1HCF
 
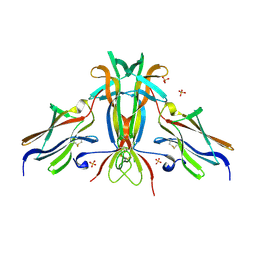 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
1CLO
 
 | |
1H5Y
 
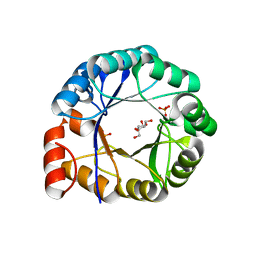 | | HisF protein from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, HISF, PHOSPHATE ION | | Authors: | Banfield, M.J, Lott, J.S, McCarthy, A.A, Baker, E.N. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AD9
 
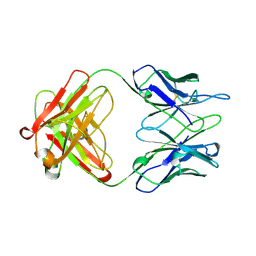 | | IGG-FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY CTM01 | | Descriptor: | IGG CTM01 FAB (HEAVY CHAIN), IGG CTM01 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-24 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1BD9
 
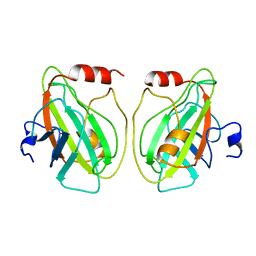 | |
1BEH
 
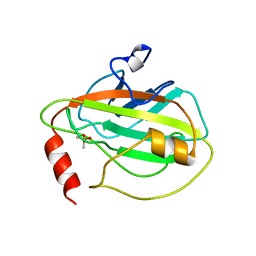 | | HUMAN PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN IN COMPLEX WITH CACODYLATE | | Descriptor: | CACODYLATE ION, PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN | | Authors: | Banfield, M.J, Barker, J.J, Perry, A, Brady, R.L. | | Deposit date: | 1998-05-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction.
Structure, 6, 1998
|
|
1AE6
 
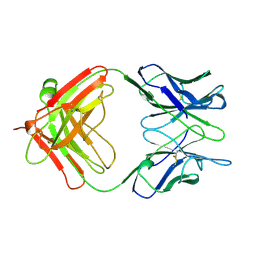 | | IGG-FAB FRAGMENT OF MOUSE MONOCLONAL ANTIBODY CTM01 | | Descriptor: | IGG CTM01 FAB (HEAVY CHAIN), IGG CTM01 FAB (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-03-06 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1AD0
 
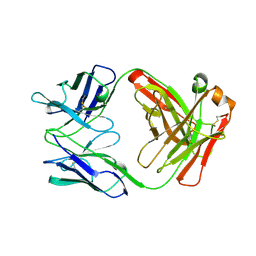 | | FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 | | Descriptor: | ANTIBODY A5B7 (HEAVY CHAIN), ANTIBODY A5B7 (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
1E3J
 
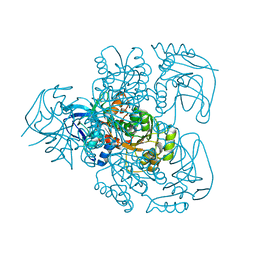 | | Ketose reductase (sorbitol dehydrogenase) from silverleaf whitefly | | Descriptor: | BORIC ACID, NADP(H)-DEPENDENT KETOSE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Banfield, M.J, Salvucci, M.E, Baker, E.N, Smith, C.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nadp(H)-Dependent Ketose Reductase from Besimia Argentifolii at 2.3 Angstrom Resolution
J.Mol.Biol., 306, 2001
|
|
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
 | |
2HX8
 
 | |
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
2FTA
 
 | |
3FN5
 
 | |
3FN6
 
 | |
3FSV
 
 | |
3FS9
 
 | |
3FT0
 
 | |
3FSW
 
 | |
3FSZ
 
 | |
