5AKQ
 
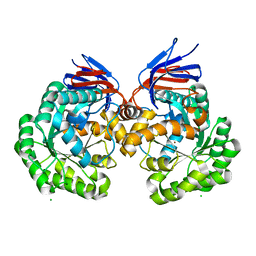 | | X-ray structure and mutagenesis studies of the N-isopropylammelide isopropylaminohydrolase, AtzC | | Descriptor: | CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ZINC ION | | Authors: | Balotra, S, Warden, A.C, Newman, J, Briggs, L.J, Scott, C, Peat, T.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CP8
 
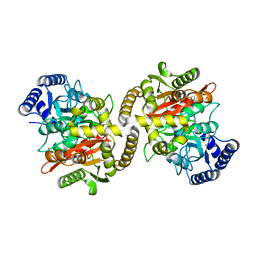 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
4CQD
 
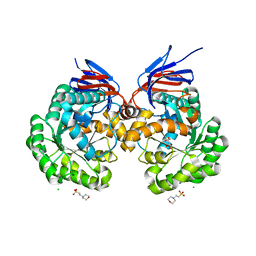 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MALONATE ION, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CQC
 
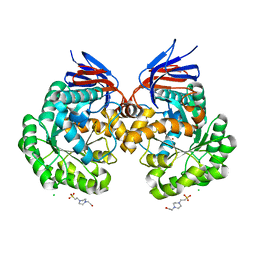 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CQB
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V1Y
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | 1,2-ETHANEDIOL, ATRAZINE CHLOROHYDROLASE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HY4
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HY0
 
 | | orotic acid hydrolase | | Descriptor: | GLYCEROL, Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HY2
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HXZ
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HY1
 
 | | high resolution structure of barbiturase | | Descriptor: | 1,3,5-triazine-2,4,6-triol, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Scott, C, Balotra, S, Wilding, M, Newman, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HXU
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, CHLORIDE ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
4BVR
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triol, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVT
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation | | Descriptor: | BARBITURIC ACID, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVQ
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
5HWE
 
 | | high resolution structure of barbiturase | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Barbiturase, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Scott, C. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
