3ZFY
 
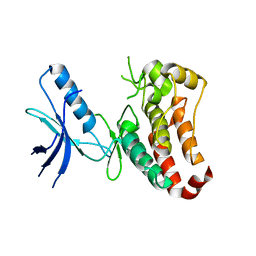 | | Crystal structure of EphB3 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 3 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZFX
 
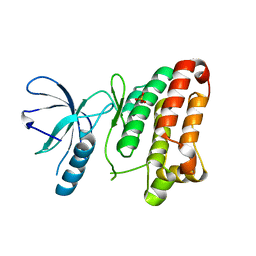 | | Crystal structure of EphB1 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 1, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZEW
 
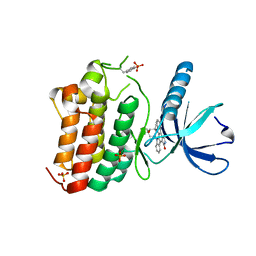 | | Crystal structure of EphB4 in complex with staurosporine | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE, SULFATE ION | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-12-25 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
3ZFM
 
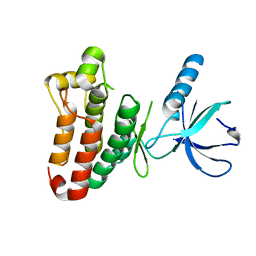 | | Crystal structure of EphB2 | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Debreczeni, J.E, Overman, R, Truman, C, McAlister, M, Attwood, T.K. | | Deposit date: | 2012-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Completing the Structural Family Portrait of the Human Ephb Tyrosine Kinase Domains
Protein Sci., 23, 2014
|
|
2YN8
 
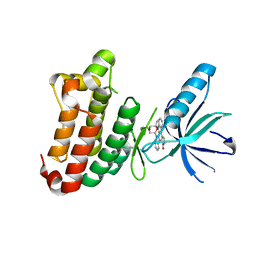 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE | | Authors: | Read, J, Brassington, C.A, Overmann, R. | | Deposit date: | 2012-10-13 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Stability and Solubility Engineering of the Ephb4 Tyrosine Kinase Catalytic Domain Using a Rationally Designed Synthetic Library.
Protein Eng.Des.Sel., 26, 2013
|
|
3NAD
 
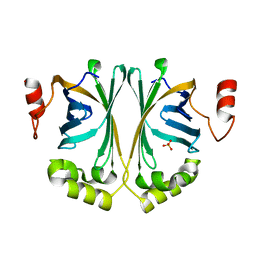 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
