2KSR
 
 | |
2LM2
 
 | | NMR structures of the transmembrane domains of the AChR b2 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit beta-2 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
8F4V
 
 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | Authors: | Bondarenko, V, Chen, Q, Tang, P. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
2LLY
 
 | | NMR structures of the transmembrane domains of the nAChR a4 subunit | | Descriptor: | Neuronal acetylcholine receptor subunit alpha-4 | | Authors: | Bondarenko, V, Mowrey, D, Tillman, T, Cui, T, Liu, L.T, Xu, Y, Tang, P. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structures of the transmembrane domains of the a4b2 nAChR.
Biochim.Biophys.Acta, 1818, 2012
|
|
2K58
 
 | |
4X22
 
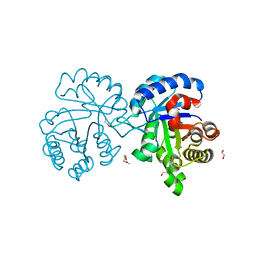 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
7RPM
 
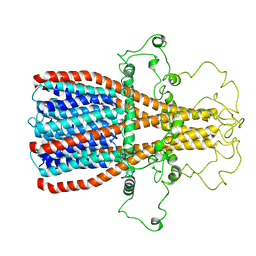 | |
4YMZ
 
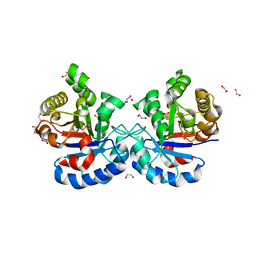 | | DHAP bound Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, SULFATE ION, ... | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-08 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
5GZP
 
 | |
4YXG
 
 | |
4YWI
 
 | | F96S/L167V Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0J
 
 | | F96S/S73A Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0S
 
 | |
1XLT
 
 | | Crystal structure of Transhydrogenase [(domain I)2:domain III] heterotrimer complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sundaresan, V, Chartron, J, Yamaguchi, M, Stout, C.D. | | Deposit date: | 2004-09-30 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational Diversity in NAD(H) and Transhydrogenase Nicotinamide Nucleotide Binding Domains
J.Mol.Biol., 346, 2005
|
|
5GV4
 
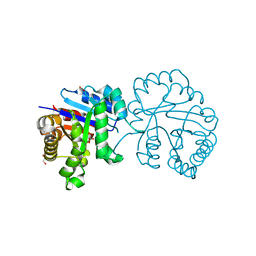 | |
1PNQ
 
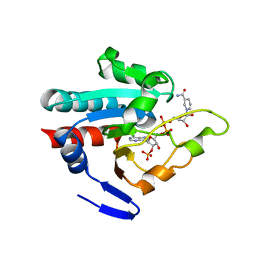 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADPH | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PNO
 
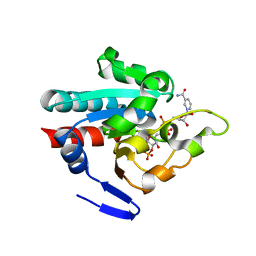 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADP | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
2MAW
 
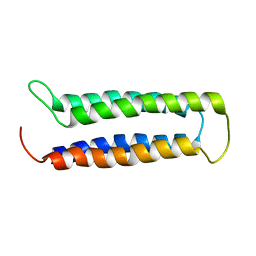 | |
2I7U
 
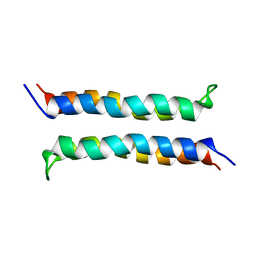 | | Structural and Dynamical Analysis of a Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets | | Descriptor: | Four-alpha-helix bundle | | Authors: | Ma, D, Brandon, N.R, Cui, T, Bondarenko, V, Canlas, C, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part I: structural and dynamical analyses.
Biophys.J., 94, 2008
|
|
8BBW
 
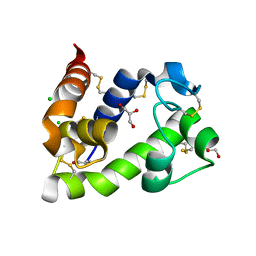 | | Crystal structure of medical leech destabilase (low salt) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for inhibition of thrombolytic destabilase
from medical leech by physiological sodium concentrations
To Be Published
|
|
5CDE
 
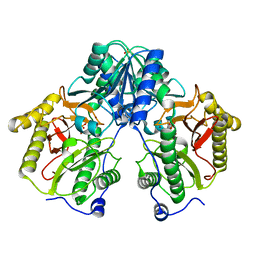 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
5CDL
 
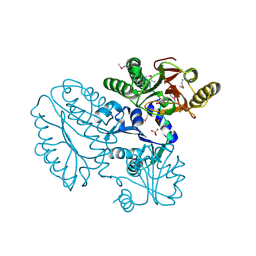 | | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-04 | | Release date: | 2016-08-10 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative)
To Be Published
|
|
5CDV
 
 | | Proline dipeptidase from Deinococcus radiodurans R1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans R1 at 1.45 Angstrom resolution
To Be Published
|
|
5CIK
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition | | Descriptor: | CITRIC ACID, GLYCEROL, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Agrawal, U, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition
To Be Published
|
|
2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
