5ZMJ
 
 | |
3A6Z
 
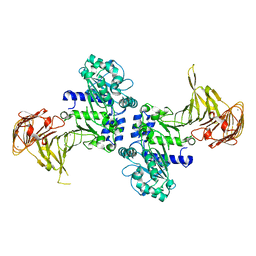 | | Crystal structure of Pseudomonas sp. MIS38 lipase (PML) in the open conformation following dialysis against Ca-free buffer | | Descriptor: | CALCIUM ION, Lipase | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-09-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
3AIL
 
 | |
3AIK
 
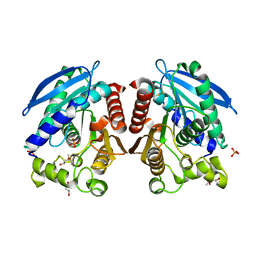 | | Crystal structure of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AIN
 
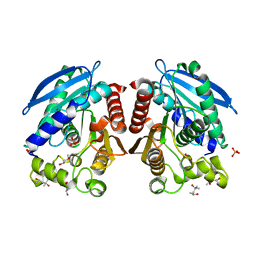 | | R267G mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AIO
 
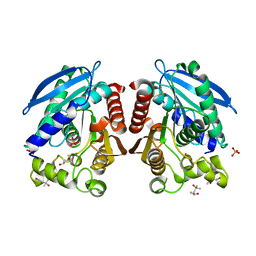 | | R267K mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
2Z8Z
 
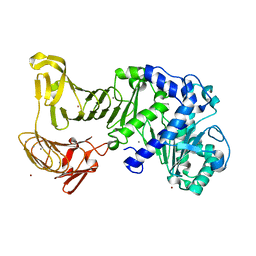 | | Crystal structure of a platinum-bound S445C mutant of Pseudomonas sp. MIS38 lipase | | Descriptor: | CALCIUM ION, Lipase, PLATINUM (II) ION, ... | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z8X
 
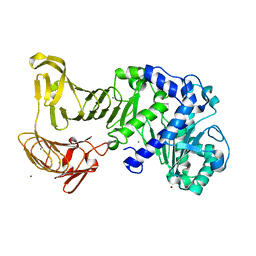 | | Crystal structure of extracellular lipase from Pseudomonas sp. MIS38 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
3A70
 
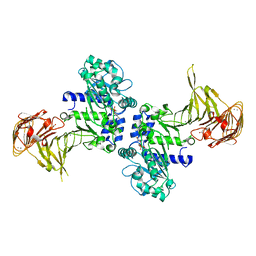 | | Crystal structure of Pseudomonas sp. MIS38 lipase in complex with diethyl phosphate | | Descriptor: | ACETATE ION, CALCIUM ION, DIETHYL PHOSPHONATE, ... | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-09-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
2ZVD
 
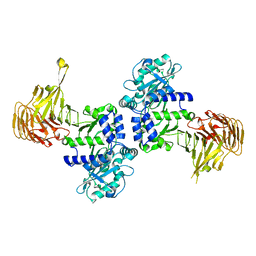 | | Crystal structure of Pseudomonas sp. MIS38 lipase in an open conformation | | Descriptor: | CALCIUM ION, Lipase | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
3AIM
 
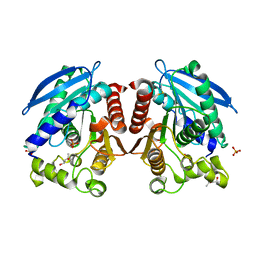 | | R267E mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3ALY
 
 | |
3AUK
 
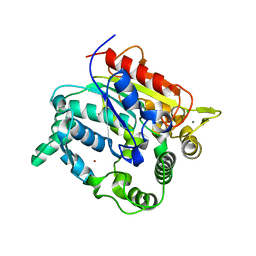 | | Crystal structure of a lipase from Geobacillus sp. SBS-4S | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lipase, ... | | Authors: | Angkawidjaja, C, Tayyab, M, Rashid, N, Kanaya, S. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a lipase from Geobacillus sp. SBS-4S
TO BE PUBLISHED
|
|
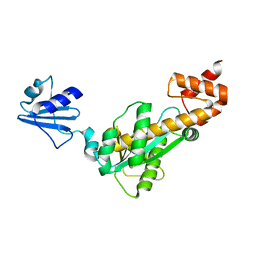
3ASM
 
 | |
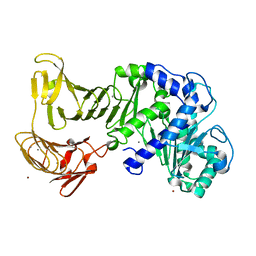
2ZJ7
 
 | |
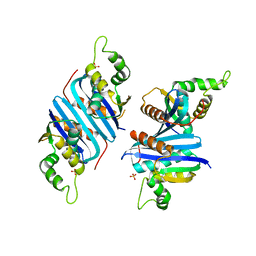
2ZQB
 
 | |
2ZJ6
 
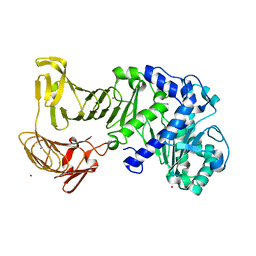 | |
5X7K
 
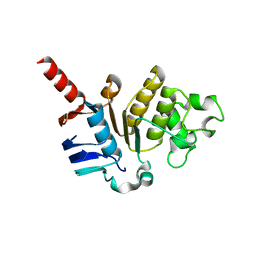 | |
4E19
 
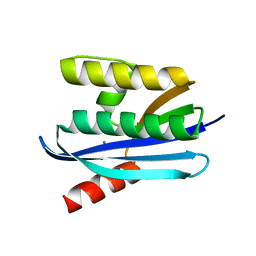 | |
4NYN
 
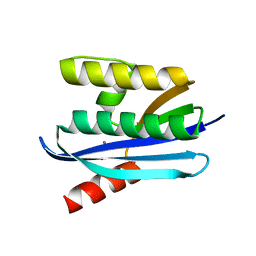 | |
3AFG
 
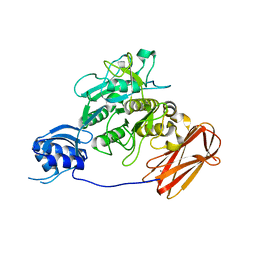 | | Crystal structure of ProN-Tk-SP from Thermococcus kodakaraensis | | Descriptor: | CALCIUM ION, Subtilisin-like serine protease | | Authors: | Foophow, T, Tanaka, S, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin homologue, Tk-SP, from Thermococcus kodakaraensis: requirement of a C-terminal beta-jelly roll domain for hyperstability.
J.Mol.Biol., 400, 2010
|
|
3AHP
 
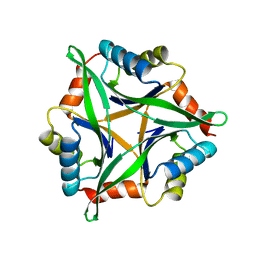 | | Crystal structure of stable protein, CutA1, from a psychrotrophic bacterium Shewanella sp. SIB1 | | Descriptor: | CutA1 | | Authors: | Tanaka, S, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-04-26 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of stable protein CutA1 from psychrotrophic bacterium Shewanella sp. SIB1
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3AOW
 
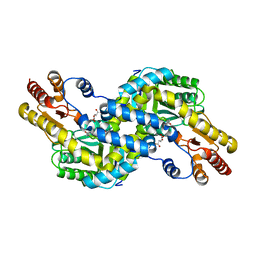 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-10-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
3AOV
 
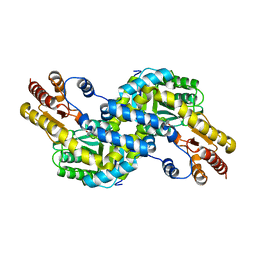 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-10-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
3B09
 
 | | Crystal structure of the N-domain of FKBP22 from Shewanella sp. SIB1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Budiman, C, Angkawidjaja, C, Motoike, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-06-07 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-domain of FKBP22 from Shewanella sp. SIB1: dimer dissociation by disruption of Val-Leu knot
Protein Sci., 20, 2011
|
|
