5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | Authors: | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | Deposit date: | 2017-07-04 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
5WCN
 
 | |
5WC8
 
 | |
5WC6
 
 | |
1KN9
 
 | |
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
6XFK
 
 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | Descriptor: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFU
 
 | |
6UEX
 
 | | Crystal structure of S. aureus LcpA in complex with octaprenyl-pyrophosphate-GlcNAc | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-hydroxy{[(S)-hydroxy{[(2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]oxy}phosphoryl]oxy}phosphoryl]-alpha-D-glucopyranose, GLYCEROL, Regulatory protein MsrR, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
7JLR
 
 | |
7JLM
 
 | |
7JLI
 
 | | Crystal structure of Bacillus subtilis UppS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Isoprenyl transferase | | Authors: | Workman, S.D, Strynadka, N.C.J. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Inhibition of Undecaprenyl Pyrophosphate Synthase from Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
7JLJ
 
 | |
6UF6
 
 | | Crystal structure of B. subtilis TagU | | Descriptor: | GLYCEROL, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagU, SULFATE ION | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF3
 
 | | Crystal structure of B. subtilis TagV | | Descriptor: | Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagV | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF5
 
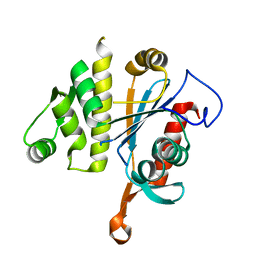 | | Crystal structure of B. subtilis TagT | | Descriptor: | Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagT | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6U2D
 
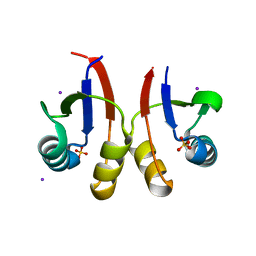 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
8V31
 
 | |
6PEP
 
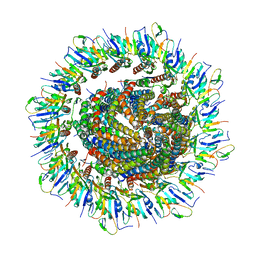 | | Focussed refinement of InvGN0N1:SpaPQR:PrgIJ from the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Protein InvG, Protein PrgH, Protein PrgI, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6PEE
 
 | |
1Q16
 
 | | Crystal structure of Nitrate Reductase A, NarGHI, from Escherichia coli | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Strynadka, N.C.J. | | Deposit date: | 2003-07-18 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A
Nat.Struct.Biol., 10, 2003
|
|
6PEM
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgHK from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q14
 
 | | Structure of the Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
