7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4ES2
 
 | |
4ES3
 
 | |
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
6NE7
 
 | | Structure of G810A mutant of RET protein tyrosine kinase domain. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NEC
 
 | | STRUCTURE OF RET PROTEIN TYROSINE KINASE DOMAIN IN COMPLEX WITH NINTEDANIB | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NJA
 
 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6OLU
 
 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
6O6H
 
 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
5W2J
 
 | | Crystal structure of dimeric form of mouse Glutaminase C | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Cerione, R.A, Li, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Basis of Glutaminase Activation: A KEY ENZYME THAT PROMOTES GLUTAMINE METABOLISM IN CANCER CELLS.
J. Biol. Chem., 291, 2016
|
|
4H7A
 
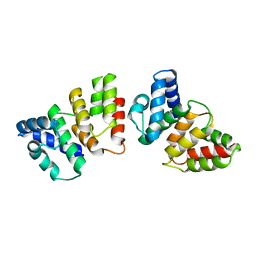 | | Crystal structure of CasB from Thermus thermophilus | | Descriptor: | CRISPR-associated protein Cse2 | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
5IC0
 
 | | Structural analysis of a talin triple domain module | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Brennan, M.L. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
4H79
 
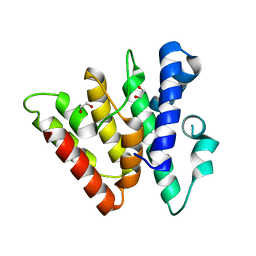 | | Crystal structure of CasB from Thermobifida fusca | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein, Cse2 family | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
6KHY
 
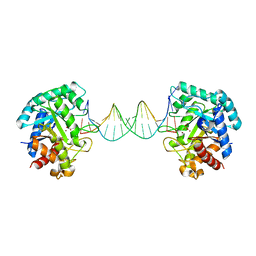 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
7E9O
 
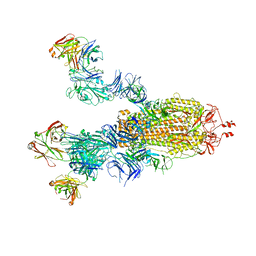 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
