8IX1
 
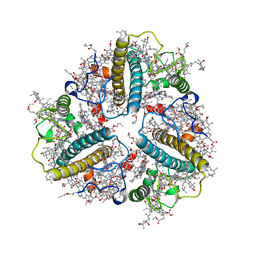 | | Cryo-EM structure of protonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
4QOC
 
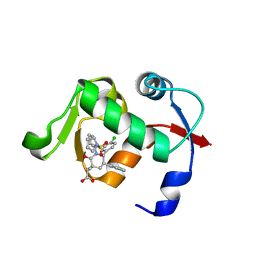 | |
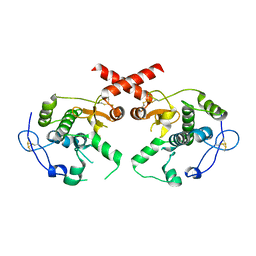
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
4L0K
 
 | |
7XAT
 
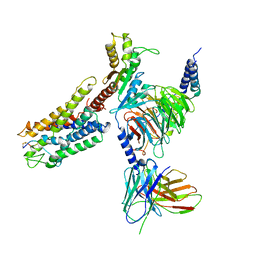 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
173L
 
 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
1D1U
 
 | | USE OF AN N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE TO FACILITATE CRYSTALLIZATION AND ANALYSIS OF A PSEUDO-16-MER DNA MOLECULE CONTAINING G-A MISPAIRS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*GP*TP*G)-3'), PROTEIN (REVERSE TRANSCRIPTASE) | | Authors: | Cote, M.L, Yohannan, S, Georgiadis, M.M. | | Deposit date: | 1999-09-21 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
149L
 
 | |
7X1T
 
 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with Taltirelin. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ScFv16, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
7X1U
 
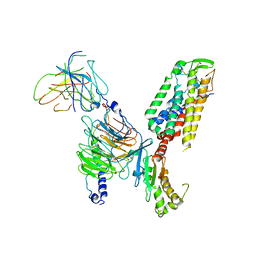 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with an Endogenous Peptide Agonist TRH. | | Descriptor: | Endogenous Peptide Agonist TRH, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
7VT0
 
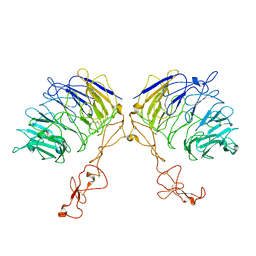 | | Dimer structure of SORLA | | Descriptor: | Sortilin-related receptor | | Authors: | Xi, Z, Cang, W, Chuang, L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
7XHA
 
 | |
7XHB
 
 | |
