5QNI
 
 | |
5QNY
 
 | |
5QOF
 
 | |
5QND
 
 | |
5QNT
 
 | |
5QO9
 
 | |
5E89
 
 | | Crystal structure of Human galectin-3 CRD in complex with 3-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
5E8A
 
 | | Crystal structure of Human galectin-3 CRD in complex with 4-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
5E88
 
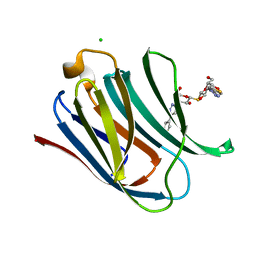 | | Crystal structure of Human galectin-3 CRD in complex with thienyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(thiophen-3-yl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-1-thio-3-[4-(thiophen-3-yl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
5T3M
 
 | |
8QPP
 
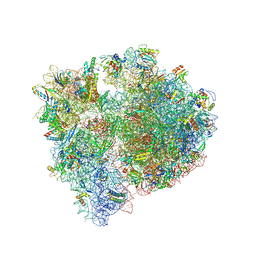 | | Bacillus subtilis MutS2-collided disome complex (stalled 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
