7WIS
 
 | |
7WIR
 
 | |
1J1D
 
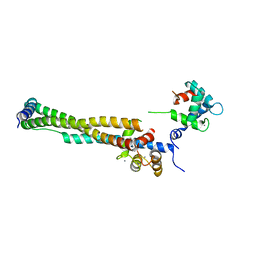 | | Crystal structure of the 46kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
1J1E
 
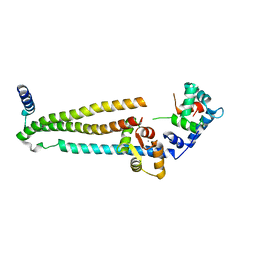 | | Crystal structure of the 52kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
2ZL8
 
 | |
2AY5
 
 | |
2AY7
 
 | |
2AY2
 
 | |
2AY3
 
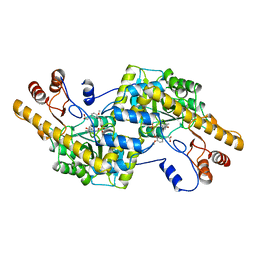 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITH 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID | | Descriptor: | 3-(3,4-DIMETHOXYPHENYL)PROPIONIC ACID, AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1998-08-06 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of Paracoccus denitrificans aromatic amino acid aminotransferase has contrary properties: flexibility and rigidity.
Biochemistry, 38, 1999
|
|
2AY1
 
 | |
2AY6
 
 | |
2AY4
 
 | |
2AY9
 
 | |
2AY8
 
 | |
1UIM
 
 | |
1UIN
 
 | |
1V2D
 
 | |
1V7C
 
 | |
1VB3
 
 | |
1V2F
 
 | |
1V2E
 
 | |
1WYC
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, DN mutant | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
1X2A
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-D-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1X28
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1X29
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-2-methyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-2-METHYL-L-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
