6DMS
 
 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
3ECQ
 
 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
6TBJ
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{S},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBH
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-[ethyl(methyl)amino]-~{N}-[6-[[(1~{S},2~{R},3~{S},4~{R})-2-(hydroxymethyl)-3,4-bis(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBG
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[[(1~{R},2~{R},3~{S},4~{S},5~{S})-2-(hydroxymethyl)-3,4,5-tris(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
1JYX
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-Galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
1JYV
 
 | |
9GPB
 
 | |
1CXK
 
 | | COMPLEX BETWEEN A MALTONONAOSE SUBSTRATE AND BACILLUS CIRCULANS STRAIN 251 CGTASE E257Q/D229N | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-02-24 | | Release date: | 1999-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family.
Nat.Struct.Biol., 6, 1999
|
|
1CXL
 
 | |
3VPL
 
 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
6TBK
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{R},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBF
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-5-(octylamino)cyclopentane-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBI
 
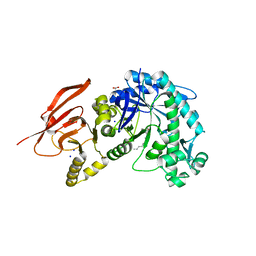 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,6R)-4-(hydroxymethyl)-6-(octylamino)cyclohex-4-ene-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
5EIY
 
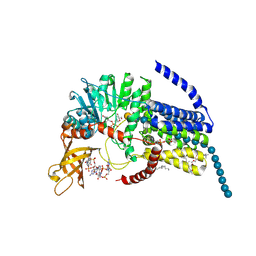 | | Bacterial cellulose synthase bound to a substrate analogue | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIUNDECYL PHOSPHATIDYL CHOLINE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | McNamara, J.T, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
5EJZ
 
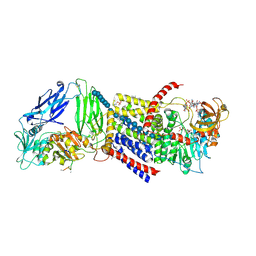 | | Bacterial Cellulose Synthase Product-Bound State | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-2-fluoro-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
5EMY
 
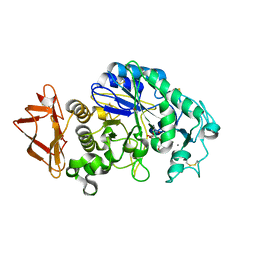 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-11-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
5EJ1
 
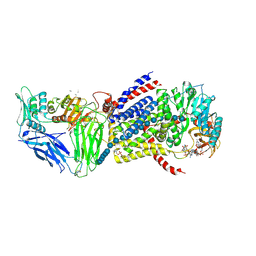 | | Pre-translocation state of bacterial cellulose synthase | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Moragn, J.L.W, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
4X6L
 
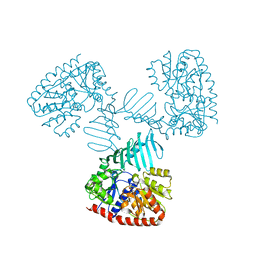 | | Crystal structure of S. aureus TarM in complex with UDP | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7P
 
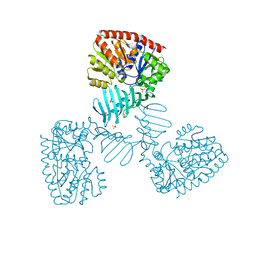 | | Crystal structure of apo S. aureus TarM | | Descriptor: | SULFATE ION, TarM | | Authors: | Worrall, L.J, Sobhanifar, S, Gruninger, R.J, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X7M
 
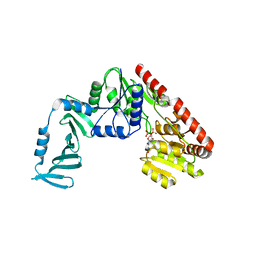 | | Crystal structure of S. aureus TarM G117R mutant in complex with UDP and UDP-GlcNAc | | Descriptor: | TarM, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, Strynadka, N.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6DLH
 
 | | Endo-fucoidan hydrolase MfFcnA4 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION, ... | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
7ZKC
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (apo form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | Deposit date: | 2022-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
