7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
8CY5
 
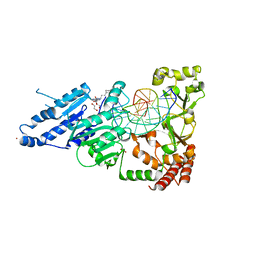 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
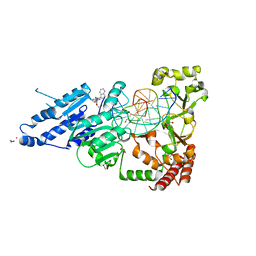 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
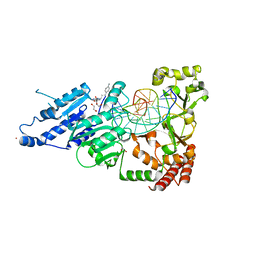 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXS
 
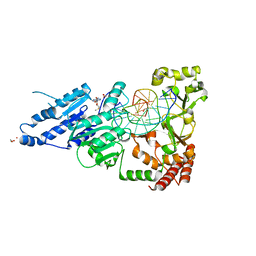 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DNA Strand 1, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXV
 
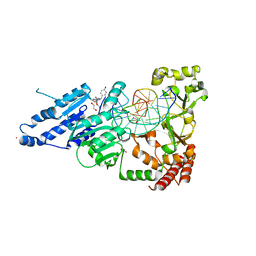 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXY
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(2-Phenethyl)adenosine (Compound 8) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXU
 
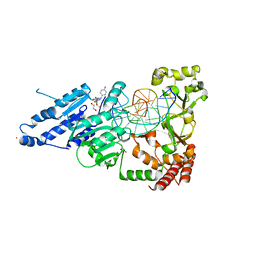 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY2
 
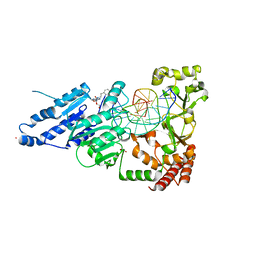 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor APNEA (Compound 9) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXT
 
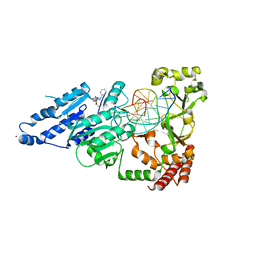 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-benzyladenosine (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXW
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor piclidenoson (Compound 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
1OBM
 
 | |
6UKG
 
 | | HhaI endonuclease in Complex With DNA in space group P21 (pH 4.2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*AP*GP*CP*GP*CP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*GP*CP*GP*CP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2019-10-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Nucleic Acids Res., 48, 2020
|
|
6UKF
 
 | |
6UKI
 
 | |
4N90
 
 | |
6UKE
 
 | |
5KE6
 
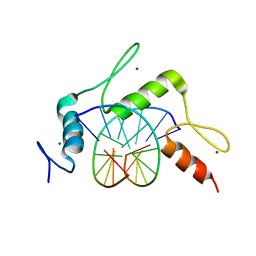 | | mouse Klf4 ZnF1-3 and TpG/CpA sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*AP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5KEB
 
 | |
