8XVL
 
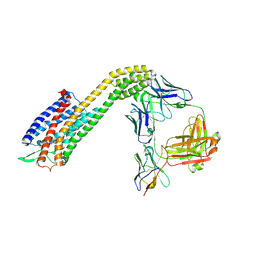 | | Cryo-EM structure of ETAR bound with Zibotentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, Zibotentan, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVI
 
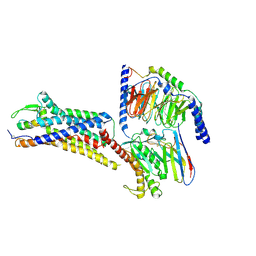 | | Cryo-EM structure of ETAR bound with Endothelin1 | | Descriptor: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
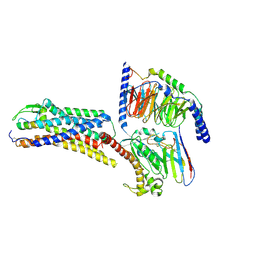 | | Cryo-EM structure of ETBR bound with Endothelin1 | | Descriptor: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
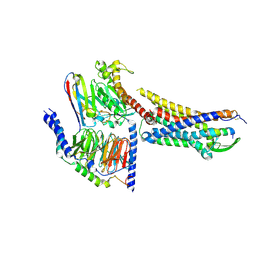 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
7X7R
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7VOO
 
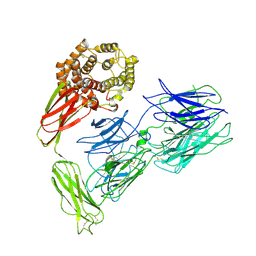 | | Induced alpha-2-macroglobulin monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Huang, X, Wang, Y, Ping, Z. | | Deposit date: | 2021-10-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the dynamic transformation of human alpha-2-macroglobulin working as a protease inhibitor.
Sci China Life Sci, 65, 2022
|
|
7D64
 
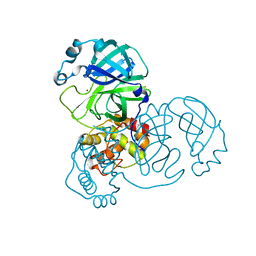 | |
6K1P
 
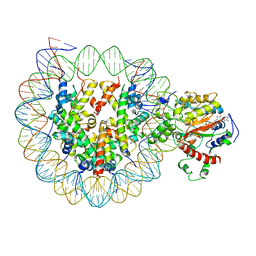 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6MWY
 
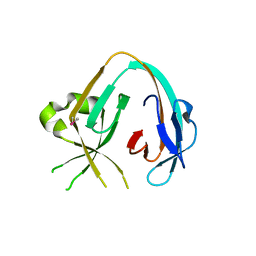 | | The Prp8 intein of Cryptococcus gattii | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Fu, B, Green, C.M, Lang, Y, Zhang, J, Oven, T.S, Li, X, Callahan, B.P, Chaturvedi, S, Belfort, M, Liao, G, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
5Z3R
 
 | | Crystal Structure of Delta 5-3-Ketosteroid Isomerase from Mycobacterium sp. | | Descriptor: | Steroid delta-isomerase | | Authors: | Cheng, X.Y, Peng, F, Yang, F, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-based reconstruction of a Mycobacterium hypothetical protein into an active Delta5-3-ketosteroid isomerase.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
8J1Z
 
 | | The global structure of pre50S related to DbpA in state3 | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yu, T, Zeng, F. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PTC Remodeling in Pre50S Intermediates: Insights into the Role of DEAD-box RNA Helicase DbpA
To Be Published
|
|
5ZB5
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
5ZY5
 
 | | spCOMT apo structure | | Descriptor: | Probable catechol O-methyltransferase 1 | | Authors: | Wang, Q, Xu, L. | | Deposit date: | 2018-05-22 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | structural and functional investigations of spCOMT
To Be Published
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY9
 
 | | tail proteins | | Descriptor: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY6
 
 | | The portal protein (GP8) of bacteriophage T7 | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
