2QOL
 
 | | Human EphA3 kinase and juxtamembrane region, Y596:Y602:S768G triple mutant | | Descriptor: | Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2RAN
 
 | | RAT ANNEXIN V CRYSTAL STRUCTURE: CA2+-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Concha, N.O, Head, J.F, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1994-09-01 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Rat annexin V crystal structure: Ca(2+)-induced conformational changes.
Science, 261, 1993
|
|
7SJK
 
 | | Structure of PLS A-domain (residues 391-656) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Pls Plasmin sensitive surface protein | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-18 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSZ
 
 | | Structure of human Kv1.3 with A0194009G09 nanobodies | | Descriptor: | Nanobody A0194009G09, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSY
 
 | | Structure of human Kv1.3 (alternate conformation) | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSV
 
 | | Structure of human Kv1.3 with Fab-ShK fusion | | Descriptor: | Fab-ShK fusion, heavy chain, light chain, ... | | Authors: | Meyerson, J.R, Selvakumar, P, Smider, V, Huang, R. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
1PAA
 
 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
7SP2
 
 | | Structure of PLS A-domain (residues 391-656; 513-518 deletion mutant) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Plasmin Sensitive Protein Pls | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of PLS A-domain (residues 391-65) from Staphylococcus aureus
Not Published
|
|
1PC9
 
 | | Crystal Structure of BnSP-6, a Lys49-Phospholipase A2 | | Descriptor: | BnSP-6 | | Authors: | Magro, A.J, Soares, A.M, Giglio, J.R, Fontes, M.R.M. | | Deposit date: | 2003-05-16 | | Release date: | 2004-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of BnSP-7 and BnSP-6, two Lys49-phospholipases A(2): quaternary structure and inhibition mechanism insights.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
7TN0
 
 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
7TIO
 
 | |
7TIR
 
 | |
7TIP
 
 | |
7TIS
 
 | |
7TIQ
 
 | |
1OXA
 
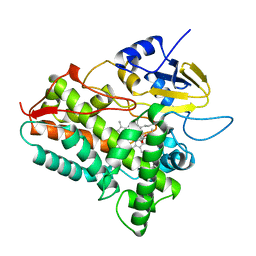 | | CYTOCHROME P450 (DONOR:O2 OXIDOREDUCTASE) | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, CYTOCHROME P450 ERYF, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 1995-07-14 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome P450eryF involved in erythromycin biosynthesis.
Nat.Struct.Biol., 2, 1995
|
|
7U4A
 
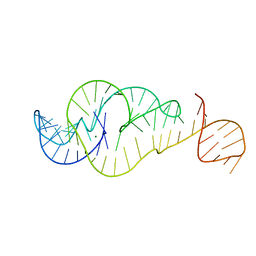 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Dynamic Basis of Xrn1 Resistance in Mosquito-borne Flavivirus RNA
To Be Published
|
|
7TVP
 
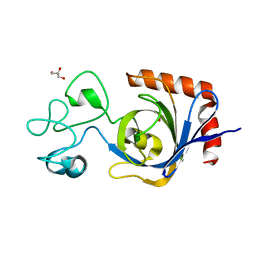 | | Viral AMG chitosanase V-Csn, E157Q mutant, chitotriose complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, GLYCEROL, Viral chitosanase V-Csn E157Q mutant chitotriose complex | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVM
 
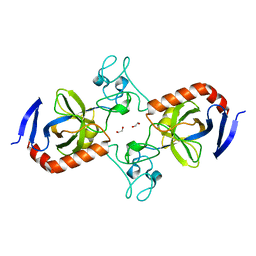 | | Viral AMG chitosanase V-Csn, apo structure, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7UYW
 
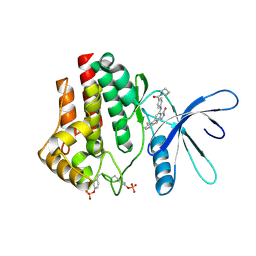 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
