4CFP
 
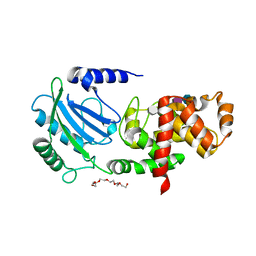 | | Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Bernardo-Garcia, N, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4CFO
 
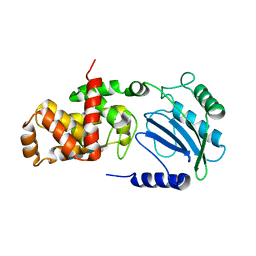 | | Structure of Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranoside, MLTC | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4C5F
 
 | |
4CHX
 
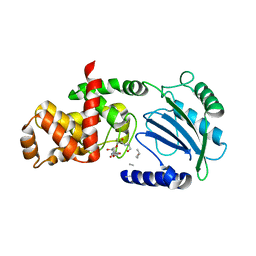 | | Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase C, ... | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-12-04 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
7PVA
 
 | | 1.9 Angstrom crystal structure of dimeric PorX, co-crystallized in the presence of zinc | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Nucleic Acids Res., 50, 2022
|
|
7PVK
 
 | | X-ray structure of dimeric PorX (T272A mutant), in complex with pGpG. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-04 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors
Nucleic Acids Res., 50, 2022
|
|
7PV7
 
 | | Crystal structure of dimeric Porphyromonas gingivalis PorX, a type 9 secretion system response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Nucleic Acids Res., 50, 2022
|
|
7QEH
 
 | | LTA-binding domain of SlpA, the S-layer protein from Lactobacillus amylovorus | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Pavkov-Keller, T. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFK
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II, Co-Crystallization with HgCl2, Mutation Ser316Cys (aa 194-362) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFG
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain III (aa 309-444) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, S-layer protein | | Authors: | Sagmeister, T, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFL
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain II (aa 199-308) | | Descriptor: | ACETATE ION, PHOSPHATE ION, S-layer protein | | Authors: | Sagmeister, T, Dordic, A, Eder, E, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLE
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I (aa 32-198) | | Descriptor: | S-layer protein | | Authors: | Sagmeister, T, Eder, M, Vejzovic, D, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLD
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I, Co-crystallization with HgCl2, Mutation Ser146Cys, (aa 32-198) | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, S-layer protein | | Authors: | Sagmeister, T, Vejzovic, D, Eder, M, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3GDD
 
 | | An inverted anthraquinone-DNA crystal structure | | Descriptor: | 5'-D(*(BRU)P*AP*GP*G)-3', MAGNESIUM ION, N,N'-(9,10-dioxo-9,10-dihydroanthracene-2,7-diyl)bis[2-(dimethylamino)acetamide] | | Authors: | Subirana, J.A, De Luchi, D, Wright, G, Gouyette, C. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a stacked anthraquinone-DNA complex
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1IJC
 
 | | Solution Structure of Bucandin, a Neurotoxin from the Venom of the Malayan Krait | | Descriptor: | bucandin | | Authors: | Torres, A.M, Kini, R.M, Nirthanan, S, Kuchel, P.W. | | Deposit date: | 2001-04-25 | | Release date: | 2001-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bucandin, a neurotoxin from the venom of the Malayan krait (Bungarus candidus).
Biochem.J., 360, 2001
|
|
8Q1O
 
 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QLG
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in closed conformation in complex with Peptide 1 | | Descriptor: | AliD, Peptide 1, ZINC ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLH
 
 | |
8QLJ
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with an unknown peptide | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLC
 
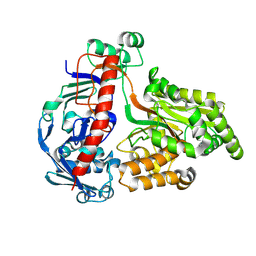 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in open conformation | | Descriptor: | AliD, MAGNESIUM ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLK
 
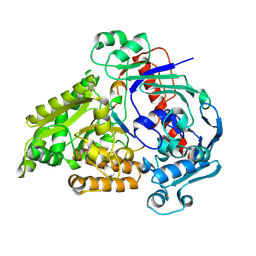 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 2 | | Descriptor: | ALA-ILE-GLN-SER-GLU-LYS-ALA-ARG-LYS-HIS-ASN, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLV
 
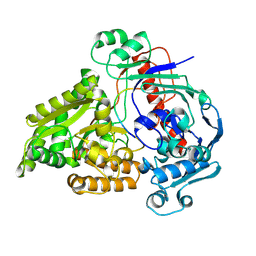 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 4 | | Descriptor: | Oligopeptide-binding protein AliB, VAL-MET-VAL-LYS-GLY-PRO-GLY-PRO-GLY-ARG | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLM
 
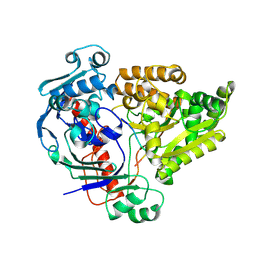 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 3 | | Descriptor: | Oligopeptide-binding protein AliB, PRO-ILE-VAL-GLY-GLY-HIS-GLU-GLY-ALA-GLY-VAL | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
