3ILD
 
 | | Structure of ORF157-K57A from Acidianus filamentous virus 1 | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
3ILE
 
 | | Crystal structure of ORF157-E86A of Acidianus filamentous virus 1 | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
4QLR
 
 | | Llama nanobody n02 raised against EAEC T6SS TssM | | Descriptor: | Llama nanobody n02 VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
3EJC
 
 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
1GPL
 
 | | RP2 LIPASE | | Descriptor: | CALCIUM ION, RP2 LIPASE | | Authors: | Withers-Martinez, C, Cambillau, C. | | Deposit date: | 1996-07-13 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig.
Structure, 4, 1996
|
|
5M30
 
 | | Structure of TssK from T6SS EAEC in complex with nanobody nb18 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Type VI secretion protein | | Authors: | Nguyen, V.S, Cambillau, C, Spinelli, C, Desmyter, A, Legrand, P, Cascales, E. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
1SLC
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
3FBL
 
 | | Crystal structure of ORF132 of the archaeal virus Acidianus Filamentous Virus 1 (AFV1) | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein | | Authors: | Goulet, A, Leulliot, N, Prangishvili, D, van Tilbeurgh, H, Campanacci, V, Cambillau, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1SLB
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
3FBZ
 
 | | Crystal structure of ORF140 of the archaeal virus Acidianus Filamentous Virus 1 (AFV1) | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Goulet, A, Prangishvili, D, van Tilbeurgh, H, Campanacci, V, Cambillau, C. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1SLA
 
 | |
3RX9
 
 | | 3D structure of SciN from an Escherichia coli Patotype | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Felisberto-Rodrigues, C, Durand, E, Aschtgen, M.-S, Blangy, S, Ortiz-Lombardia, M, Douzy, B, Cambillau, C, Cascales, E. | | Deposit date: | 2011-05-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar.
Plos Pathog., 7, 2011
|
|
2FSD
 
 | |
2H85
 
 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H8V
 
 | | Structure of empty Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Briand, L, Pernollet, J.-C, Cambillau, C, Tegoni, M. | | Deposit date: | 2006-06-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational Changes of the Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L upon Ligand Binding
To be Published
|
|
4GRW
 
 | | Structure of a complex of human IL-23 with 3 Nanobodies (Llama vHHs) | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, Nanobody 124C4, ... | | Authors: | Desmyter, A, Spinelli, S, Button, C, Saunders, M, de Haard, H, Rommelaere, H, Union, A, Cambillau, C. | | Deposit date: | 2012-08-27 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex.
Front Immunol, 8, 2017
|
|
4HEP
 
 | | Complex of lactococcal phage TP901-1 with a llama vHH (vHH17) binder (nanobody) | | Descriptor: | BPP, SULFATE ION, vHH17 domain | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2HLV
 
 | | Bovine Odorant Binding Protein deswapped triple mutant | | Descriptor: | 3,6-BIS(METHYLENE)DECANOIC ACID, GLYCEROL, Odorant-binding protein | | Authors: | Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2006-07-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deswapping bovine odorant binding protein.
Biochim.Biophys.Acta, 1784, 2008
|
|
4HV0
 
 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
4EQQ
 
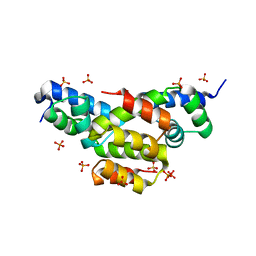 | | Structure of Ltp, a superinfection exclusion protein from the Streptococcus thermophilus temperate phage TP-J34 | | Descriptor: | PHOSPHATE ION, Putative host cell surface-exposed lipoprotein, SULFATE ION | | Authors: | Bebeacua, C, Lorenzo, C, Blangy, S, Spinelli, S, Heller, K, Cambillau, C. | | Deposit date: | 2012-04-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target.
Mol.Microbiol., 89, 2013
|
|
2X53
 
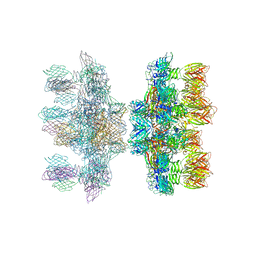 | | Structure of the phage p2 baseplate in its activated conformation with Sr | | Descriptor: | ORF15, ORF16, PUTATIVE RECEPTOR BINDING PROTEIN, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2BSE
 
 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein in complex with a llama VHH domain | | Descriptor: | LLAMA IMMUNOGLOBULIN, RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, de Haard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5LMJ
 
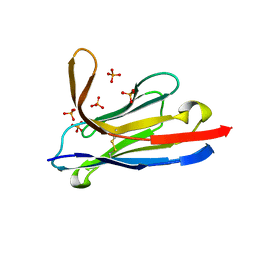 | | Llama nanobody PorM_19 | | Descriptor: | PHOSPHATE ION, nanobody | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2BSD
 
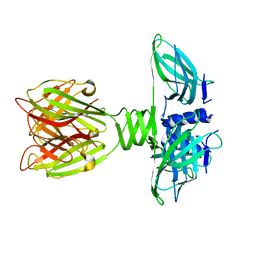 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein | | Descriptor: | RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, Dehaard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor-Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4JEI
 
 | | Nonglycosylated Yarrowia lipolytica LIP2 lipase | | Descriptor: | Lipase 2 | | Authors: | Aloulou, A, Benarouche, A, Puccinelli, D, Spinelli, S, Cavalier, J.-F, Cambillau, C, Carriere, F. | | Deposit date: | 2013-02-27 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of non-glycosylated Yarrowia lipolytica LIP2 lipase
To be published
|
|
