1K0V
 
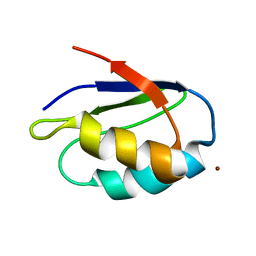 | | Copper trafficking: the solution structure of Bacillus subtilis CopZ | | Descriptor: | COPPER (I) ION, CopZ | | Authors: | Banci, L, Bertini, I, Del Conte, R, Markey, J, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Copper trafficking: the solution structure of Bacillus subtilis CopZ.
Biochemistry, 40, 2001
|
|
1JWW
 
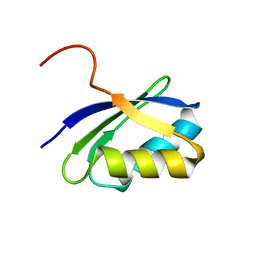 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1GIW
 
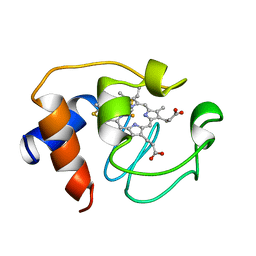 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
1J6Q
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
1K3H
 
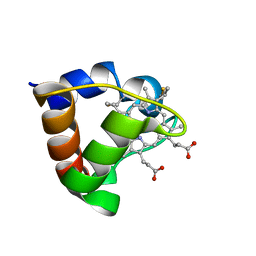 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
1KWJ
 
 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
1RK7
 
 | | Solution structure of apo Cu,Zn Superoxide Dismutase: role of metal ions in protein folding | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding
Biochemistry, 42, 2003
|
|
1S6U
 
 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1L3O
 
 | | SOLUTION STRUCTURE DETERMINATION OF THE FULLY OXIDIZED DOUBLE MUTANT K9-10A CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS, ENSEMBLE OF 35 STRUCTURES | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
1LMS
 
 | | Structural model for an alkaline form of ferricytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Dolfi, A, Turano, P, Mauk, A.G, Rosell, F.I, Gray, H.B. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural model for an alkaline form of ferricytochrome c
J.Am.Chem.Soc., 125, 2003
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SO9
 
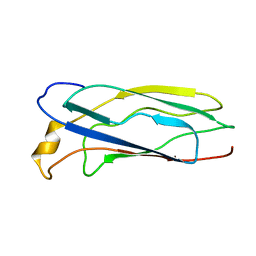 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
1S4I
 
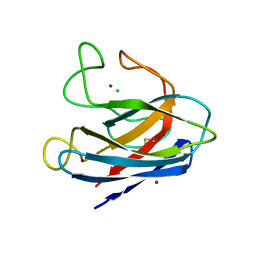 | | Crystal structure of a SOD-like protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, ZINC ION, superoxide dismutase-like protein yojM | | Authors: | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | Deposit date: | 2004-01-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1SB6
 
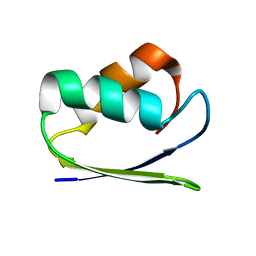 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
1MNY
 
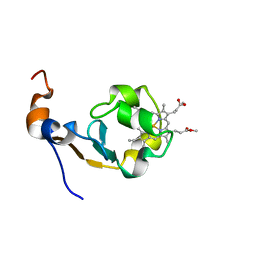 | | Dimethyl propionate ester heme-containing cytochrome b5 | | Descriptor: | DIMETHYL PROPIONATE ESTER HEME, cytochrome b5 | | Authors: | Banci, L, Bertini, I, Branchini, B.R, Hajieva, P, Spyroulias, G.A, Turano, P. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dimethyl propionate ester heme-containing cytochrome b5: structure and stability.
J.BIOL.INORG.CHEM., 6, 2001
|
|
1MWY
 
 | | Solution structure of the N-terminal domain of ZntA in the apo-form | | Descriptor: | ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1MWZ
 
 | | Solution structure of the N-terminal domain of ZntA in the Zn(II)-form | | Descriptor: | ZINC ION, ZntA | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Finney, L.A, Outten, C.E, O'Halloran, T.V. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new zinc-protein coordination site in intracellular metal trafficking: solution structure of the apo and Zn(II) forms of ZntA (46-118)
J.Mol.Biol., 323, 2002
|
|
1OQ3
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
