6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
5USH
 
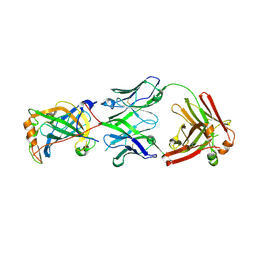 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5USL
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv304 | | Descriptor: | Fab vv304 Heavy chain, Fab vv304 Light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
6VIL
 
 | |
2WW3
 
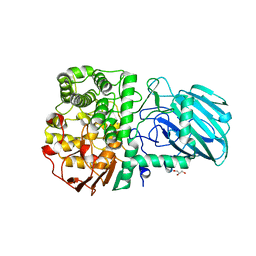 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with thiomannobioside | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Insights Into a Ca(2+)-Dependent Family of Alpha-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WW1
 
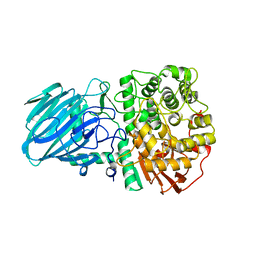 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Thiomannobioside | | Descriptor: | PUTATIVE ALPHA-1,2-MANNOSIDASE, alpha-D-mannopyranose-(1-2)-methyl 2-thio-alpha-D-mannopyranoside | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WVX
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALPHA-1,2-MANNOSIDASE | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WW0
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 1S-8AB-OCTAHYDRO-INDOLIZIDINE-1A,2A,8B-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Suits, M.D.L, Thompson, A, Zhu, Y, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
6CB6
 
 | |
6CB7
 
 | |
2WVY
 
 | | STRUCTURE OF THE FAMILY GH92 INVERTING MANNOSIDASE BT2199 FROM BACTEROIDES THETAIOTAOMICRON VPI-5482 | | Descriptor: | ALPHA-1,2-MANNOSIDASE | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WVZ
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | CALCIUM ION, GLYCEROL, KIFUNENSINE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WW2
 
 | | Structure of the Family GH92 Inverting Mannosidase BT2199 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1S-8AB-OCTAHYDRO-INDOLIZIDINE-1A,2A,8B-TRIOL, ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
4QAU
 
 | | Crystal structure of F43Y mutant of sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y, Tan, X, Li, W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | A Novel Tyrosine-Heme C-O Covalent Linkage in F43Y Myoglobin: A New Post-translational Modification of Heme Proteins
Chembiochem, 16, 2015
|
|
6BR8
 
 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BR9
 
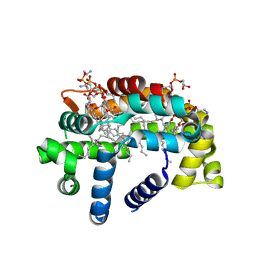 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8T1L
 
 | | Atomic model of the mammalian mouse Mediator complex with CKM module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.83 Å) | | Cite: | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
To Be Published
|
|
8T1I
 
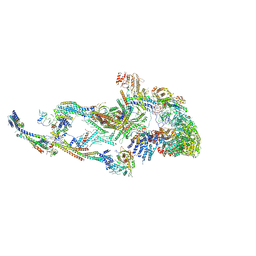 | | Atomic model of the mammalian Mediator complex with MED26 subunit | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8T9D
 
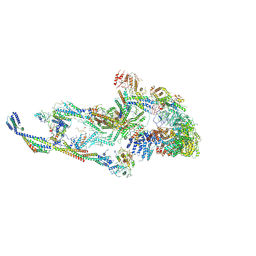 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8VES
 
 | |
8VER
 
 | |
6WIG
 
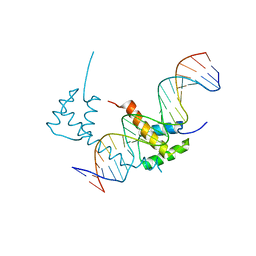 | | Structure of STENOFOLIA Protein HD domain bound with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*AP*AP*TP*AP*AP*AP*TP*CP*AP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*AP*AP*TP*GP*AP*TP*TP*TP*AP*TP*TP*CP*AP*AP*G)-3'), STENOFOLIA | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2020-04-09 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the unique tetrameric STENOFOLIA homeodomain bound with target promoter DNA.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ACG
 
 | |
6ACJ
 
 | |
6ACK
 
 | |
