5NY5
 
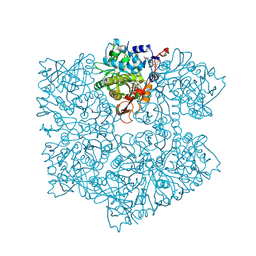 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | Descriptor: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | Authors: | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-13 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6TFU
 
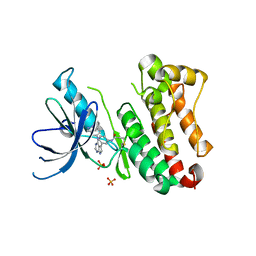 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 14d | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[3-[4-[[1-(phenylmethyl)indazol-5-yl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
6TG0
 
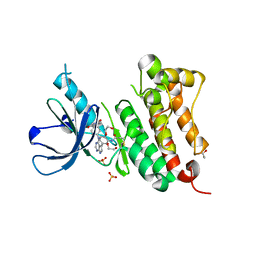 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 21a | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
1OHQ
 
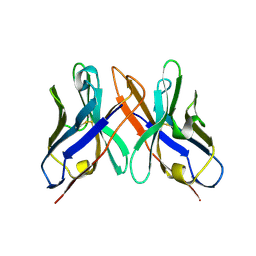 | | Crystal structure of HEL4, a soluble human VH antibody domain resistant to aggregation | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Jespers, L, Schon, O, James, L.C, Veprintsev, D, Winter, G. | | Deposit date: | 2003-05-30 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hel4, a Soluble, Refoldable Human V(H) Single Domain with a Germ-Line Scaffold
J.Mol.Biol., 337, 2004
|
|
1OM2
 
 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
1ONA
 
 | | CO-CRYSTALS OF CONCANAVALIN A WITH METHYL-3,6-DI-O-(ALPHA-D-MANNOPYRANOSYL)-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bouckaert, J, Maes, D, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1996-07-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-D-mannopyranosyl)-alpha-D-mannopyranoside reveals two binding modes.
J.Biol.Chem., 271, 1996
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
1OP8
 
 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
6TKI
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - tetragonal form at 12.7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NKE
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3a | | Descriptor: | 2-[[3-bromanyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
2J1U
 
 | |
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
6TNH
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
1OS1
 
 | | Structure of Phosphoenolpyruvate Carboxykinase complexed with ATP,Mg, Ca and pyruvate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sudom, A, Walters, R, Pastushok, L, Goldie, D, Prasad, L, Delbaere, L.T, Goldie, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin.
J.BACTERIOL., 185, 2003
|
|
6TNT
 
 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Barford, D, Yatskevich, S. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
2IL5
 
 | |
1OUS
 
 | | Lecb (PA-LII) calcium-free | | Descriptor: | SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVS
 
 | | LecB (PA-LII) in complex with core trimannoside | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
2IJ2
 
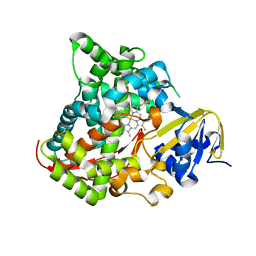 | | Atomic structure of the heme domain of flavocytochrome P450-BM3 | | Descriptor: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Helen, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
6T03
 
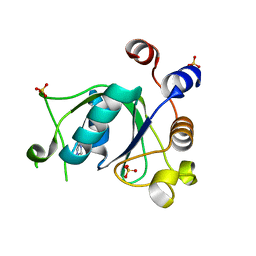 | | Crystal structure of YTHDC1 with fragment 16 (DHU_DC1_017) | | Descriptor: | 1,3-dihydroimidazole-2-thione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T1J
 
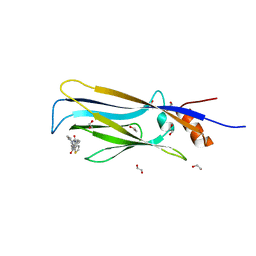 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
5N5C
 
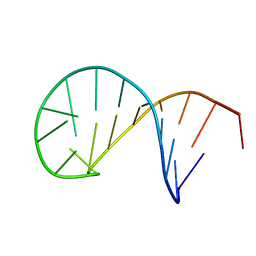 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
6SK5
 
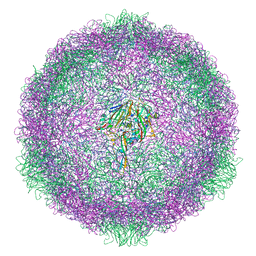 | | Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340 | | Descriptor: | 6-phenyl-~{N}3-[4-(trifluoromethyl)phenyl]-1~{H}-pyrazolo[3,4-d]pyrimidine-3,4-diamine, Rhinovirus B5 VP1, Rhinovirus B5 VP2, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5NF8
 
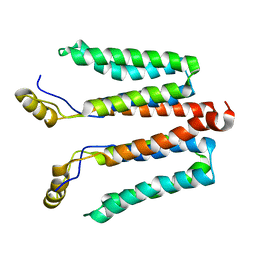 | | Solution structure of detergent-solubilized Rcf1, a yeast mitochondrial inner membrane protein involved in respiratory Complex III/IV supercomplex formation | | Descriptor: | Respiratory supercomplex factor 1, mitochondrial | | Authors: | Zhou, S, Pettersson, P, Sjoholm, J, Sjostrand, D, Hogbom, M, Brzezinski, P, Maler, L, Adelroth, P. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast Rcf1, a protein involved in respiratory supercomplex formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
