7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5M
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 1.8 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
7VTY
 
 | | de novo designed protein | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-31 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQV
 
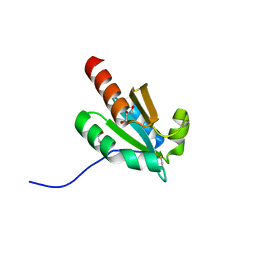 | | de novo design based on 1r26 | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VU4
 
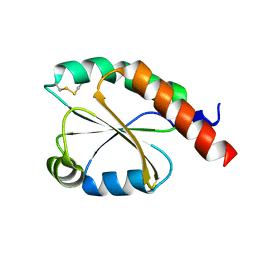 | | de novo design based on 1r26 | | Descriptor: | de novo design protein | | Authors: | Zhang, L. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQL
 
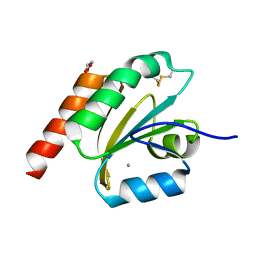 | | de novo designed based on 1r26 | | Descriptor: | AMMONIUM ION, GLYCEROL, de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQW
 
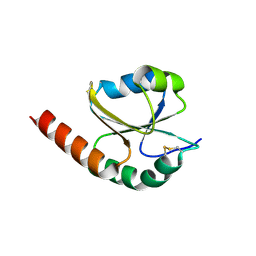 | | de novo designed protein based on 1r26 | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7C5F
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.88 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization and structure of glyceraldehyde-3-phosphate dehydrogenase type 1 from Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5N5O
 
 | |
5NH0
 
 | |
7BX8
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
5YVF
 
 | | Crystal structure of BFA1 | | Descriptor: | BFA1 | | Authors: | Pu, H, Zhang, L, Duan, Z.K, Peng, L.W, Liu, L. | | Deposit date: | 2017-11-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1.
Plant Cell, 30, 2018
|
|
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
4RHD
 
 | | DNA Duplex with Novel ZP Base Pair | | Descriptor: | DNA 9mer novel P nucleobase, DNA 9mer novel Z nucleobase, MAGNESIUM ION | | Authors: | Zhang, W, Zhang, L, Benner, S, Huang, Z. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of functional six-nucleotide DNA.
J.Am.Chem.Soc., 137, 2015
|
|
7BIP
 
 | | Crystal structure of monooxygenase RslO1 from Streptomyces bottropensis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhang, L, Zuo, C, Bechthold, A, Einsle, O. | | Deposit date: | 2021-01-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biosynthesis of the Tricyclic Aromatic Type II Polyketide Rishirilide: New Potential Third Ring Oxygenation after Three Cyclization Steps.
Mol Biotechnol., 63, 2021
|
|
7BIO
 
 | | Crystal structure of monooxygenase RslO4 from Streptomyces bottropensis | | Descriptor: | ACETATE ION, ETHANOL, Monooxygenase/putative anthronoxygenase, ... | | Authors: | Zhang, L, Zuo, C, Alali, A, Bechthold, A, Einsle, O. | | Deposit date: | 2021-01-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Biosynthesis of the Tricyclic Aromatic Type II Polyketide Rishirilide: New Potential Third Ring Oxygenation after Three Cyclization Steps.
Mol Biotechnol., 63, 2021
|
|
5B0J
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0M
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-dodecyl maltoside | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0K
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-decyl maltoside | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B01
 
 | |
5B0I
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-octyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, octyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B03
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
